-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy path1.4-Use_Cases.qmd
120 lines (69 loc) · 2.53 KB
/
1.4-Use_Cases.qmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
---
title: "Use Cases"
subtitle: ""
author:
- Elizabeth King
- Kevin Middleton
format:
revealjs:
theme: [default, custom.scss]
standalone: true
self-contained: true
logo: QMLS_Logo.png
slide-number: true
show-slide-number: all
---
## Potential & Limitations of Monte Carlo Methods
```{r}
#| label: setup
#| echo: false
#| warning: false
#| message: false
library(tidyverse)
library(cowplot)
ggplot2::theme_set(theme_cowplot())
```
- Highly flexible
- Few assumptions
- Need to demonstrate validity for new applications
## Case Studies
{width=100%}
> Taken directly from our own experiences
## Automated counting of fruit fly eggs
{width=60% fig-align="center"}
## Automated counting of fruit fly eggs
{width=80%}
## Turning flight in hummingbirds
{fig-align="center" width=100%}
## Turning flight in hummingbirds
{fig-align="center" width=100%}
## Turning flight in hummingbirds
{fig-align="center" width=100%}
## Turning flight in hummingbirds
{fig-align="center" width=100%}
## Mosaic evolution in Mesozoic birds
{fig-align="center" width=100%}
## Mosaic evolution in Mesozoic birds
{fig-align="center" width=80%}
## Mosaic evolution in Mesozoic birds
{fig-align="center" width=70%}
## Candidate Genetic Loci in the DSPR {.smaller}
- Power analysis via simulation of genetic mapping (see Quant Methods 1, 11-3)
- Detecting multiallelism
- Simulating a null for differences between allele frequencies
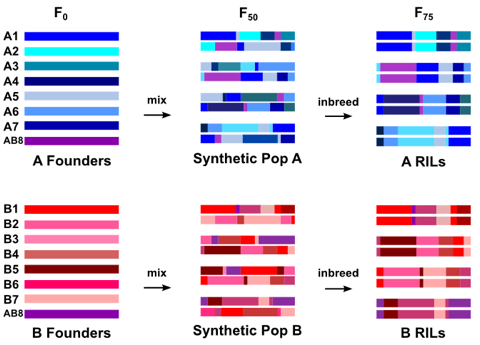{fig-align="center" width=80%}
## Multiallelism
{width=60% fig-align="center"}
## Multiallelism
{width=55% fig-align="center"}
## Allele Frequency Differences
{width=90% fig-align="center"}
## Allele Frequency Differences
{width=100% fig-align="center"}
## What questions do we ask when we use statistics? {.smaller}
1. Parameter (point) estimation
- Given a model, with unknown parameters ($\theta_0$, $\theta_1$, ..., $\theta_k$), how to estimate values of those parameters?
2. Interval estimation
- How to quantify the uncertainty associated with parameter estimates?
3. Hypothesis testing
- How to test hypotheses about parameter estimates?