diff --git a/.gitignore b/.gitignore
index 6e6ddae..2c94a80 100644
--- a/.gitignore
+++ b/.gitignore
@@ -1,2 +1,3 @@
*.npz
*.pkl
+*.pyc
diff --git a/README.md b/README.md
index 6a7498f..e69de29 100644
--- a/README.md
+++ b/README.md
@@ -1,188 +0,0 @@
-# SalGAN: Visual Saliency Prediction with Generative Adversarial Networks
-
-| ![Junting Pan][JuntingPan-photo] | ![Cristian Canton Ferrer][CristianCanton-photo] | ![Kevin McGuinness][KevinMcGuinness-photo] | ![Noel O'Connor][NoelOConnor-photo] | ![Jordi Torres][JordiTorres-photo] |![Elisa Sayrol][ElisaSayrol-photo] | ![Xavier Giro-i-Nieto][XavierGiro-photo] |
-|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
-| [Junting Pan][JuntingPan-web] | [Cristian Canton Ferrer][CristianCanton-web] | [Kevin McGuinness][KevinMcGuinness-web] | [Noel O'Connor][NoelOConnor-web] | [Jordi Torres][JordiTorres-web] | [Elisa Sayrol][ElisaSayrol-web] | [Xavier Giro-i-Nieto][XavierGiro-web] |
-
-[JuntingPan-web]: https://www.linkedin.com/in/junting-pan
-[CristianCanton-web]: https://cristiancanton.github.io/
-[KevinMcGuinness-web]: https://www.insight-centre.org/users/kevin-mcguinness
-[JordiTorres-web]: jorditorres.org
-[ElisaSayrol-web]: https://imatge.upc.edu/web/people/elisa-sayrol
-[NoelOConnor-web]: https://www.insight-centre.org/users/noel-oconnor
-[XavierGiro-web]: https://imatge.upc.edu/web/people/xavier-giro
-
-[JuntingPan-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/JuntingPan.jpg "Junting Pan"
-[KevinMcGuinness-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/authors/Kevin160x160%202.jpg?token=AFOjyZmLlX3ZgpkNe60Vn3ruTsq01rD9ks5YdAaiwA%3D%3D "Kevin McGuinness"
-[CristianCanton-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/authors/CristianCanton.jpg?token=AFOjyS9qMOnUPVLZpqN80ChO0R-x0SI5ks5Yc3qJwA%3D%3D "Cristian Canton"
-[JordiTorres-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/authors/JordiTorres.jpg?token=AFOjyUaOhEyX2MGayU2C4tExpQeT0jFUks5Yc3vcwA%3D%3D
-[ElisaSayrol-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/ElisaSayrol.jpg "Elisa Sayrol"
-[NoelOConnor-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/NoelOConnor.jpg "Noel O'Connor"
-[XavierGiro-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/XavierGiro.jpg "Xavier Giro-i-Nieto"
-
-A joint collaboration between:
-
-| ![logo-insight] | ![logo-dcu] | ![logo-microsoft] |![logo-bsc] | ![logo-gpi] |
-|:-:|:-:|:-:|:-:|:-:|
-| [Insight Centre for Data Analytics][insight-web] | [Dublin City University (DCU)][dcu-web] | [Microsoft][microsoft-web]|[Barcelona Supercomputing Center][bsc-web] | [UPC Image Processing Group][gpi-web] |
-
-[insight-web]: https://www.insight-centre.org/
-[dcu-web]: http://www.dcu.ie/
-[microsoft-web]: https://www.microsoft.com/en-us/research/
-[bsc-web]: https://www.bsc.es/
-[upc-web]: http://www.upc.edu/?set_language=en
-[etsetb-web]: https://www.etsetb.upc.edu/en/
-[gpi-web]: https://imatge.upc.edu/web/
-
-
-[logo-insight]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/insight.jpg "Insight Centre for Data Analytics"
-[logo-dcu]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/dcu.png "Dublin City University"
-[logo-microsoft]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/logos/microsoft.jpg?token=AFOjyc8Q1kkjcWIP-yen0FTEo0lsWPk6ks5Yc3j4wA%3D%3D "Microsoft"
-[logo-bsc]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/logos/bsc320x86.jpg?token=AFOjyWSHWWVvzTXnYh1DiFvH2VoWykA3ks5Yc6Q1wA%3D%3D
-[logo-upc]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/upc.jpg "Universitat Politecnica de Catalunya"
-[logo-etsetb]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/etsetb.png "ETSETB TelecomBCN"
-[logo-gpi]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/gpi.png "UPC Image Processing Group"
-
-
-## Abstract
-
-We introduce SalGAN, a deep convolutional neural network for visual saliency prediction trained with adversarial examples.
-The first stage of the network consists of a generator model whose weights are learned by back-propagation computed from a binary cross entropy (BCE) loss over downsampled versions of the saliency maps. The resulting prediction is processed by a discriminator network trained to solve a binary classification task between the saliency maps generated by the generative stage and the ground truth ones. Our experiments show how adversarial training allows reaching state-of-the-art performance across different metrics when combined with a widely-used loss function like BCE.
-
-## Slides
-
-
-
-
-
-## Publication
-
-Find the pre-print version of our work on [arXiv](https://arxiv.org/abs/1701.01081).
-
-
-
-Please cite with the following Bibtex code:
-
-```
-@InProceedings{Pan_2017_SalGAN,
-author = {Pan, Junting and Canton, Cristian and McGuinness, Kevin and O'Connor, Noel E. and Torres, Jordi and Sayrol, Elisa and Giro-i-Nieto, Xavier and},
-title = {SalGAN: Visual Saliency Prediction with Generative Adversarial Networks},
-booktitle = {arXiv},
-month = {January},
-year = {2017}
-}
-```
-
-You may also want to refer to our publication with the more human-friendly Chicago style:
-
-*Junting Pan, Cristian Canton, Kevin McGuinness, Noel E. O'Connor, Jordi Torres, Elisa Sayrol and Xavier Giro-i-Nieto. "SalGAN: Visual Saliency Prediction with Generative Adversarial Networks." arXiv. 2017.*
-
-
-
-## Models
-
-The SalGAN presented in our work can be downloaded from the links provided below the figure:
-
-SalGAN Architecture
-![architecture-fig]
-
-* [[SalGAN Generator Model (127 MB)]](https://imatge.upc.edu/web/sites/default/files/resources/1720/saliency/2017-salgan/gen_modelWeights0090.npz)
-* [[SalGAN Discriminator (3.4 MB)]](https://imatge.upc.edu/web/sites/default/files/resources/1720/saliency/2017-salgan/discrim_modelWeights0090.npz)
-
-[architecture-fig]: https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/junting/figs/fullarchitecture.jpg?token=AFOjyaH8cuBFWpldWWzo_TKVB-zekfxrks5Yc4NQwA%3D%3D "SALGAN architecture"
-[shallow-model]: https://imatge.upc.edu/web/sites/default/files/resources/1720/saliency/2016-cvpr/shallow_net.pickle
-[deep-model]: https://imatge.upc.edu/web/sites/default/files/resources/1720/saliency/2016-cvpr/deep_net_model.caffemodel
-[deep-prototxt]: https://imatge.upc.edu/web/sites/default/files/resources/1720/saliency/2016-cvpr/deep_net_deploy.prototxt
-
-## Visual Results
-
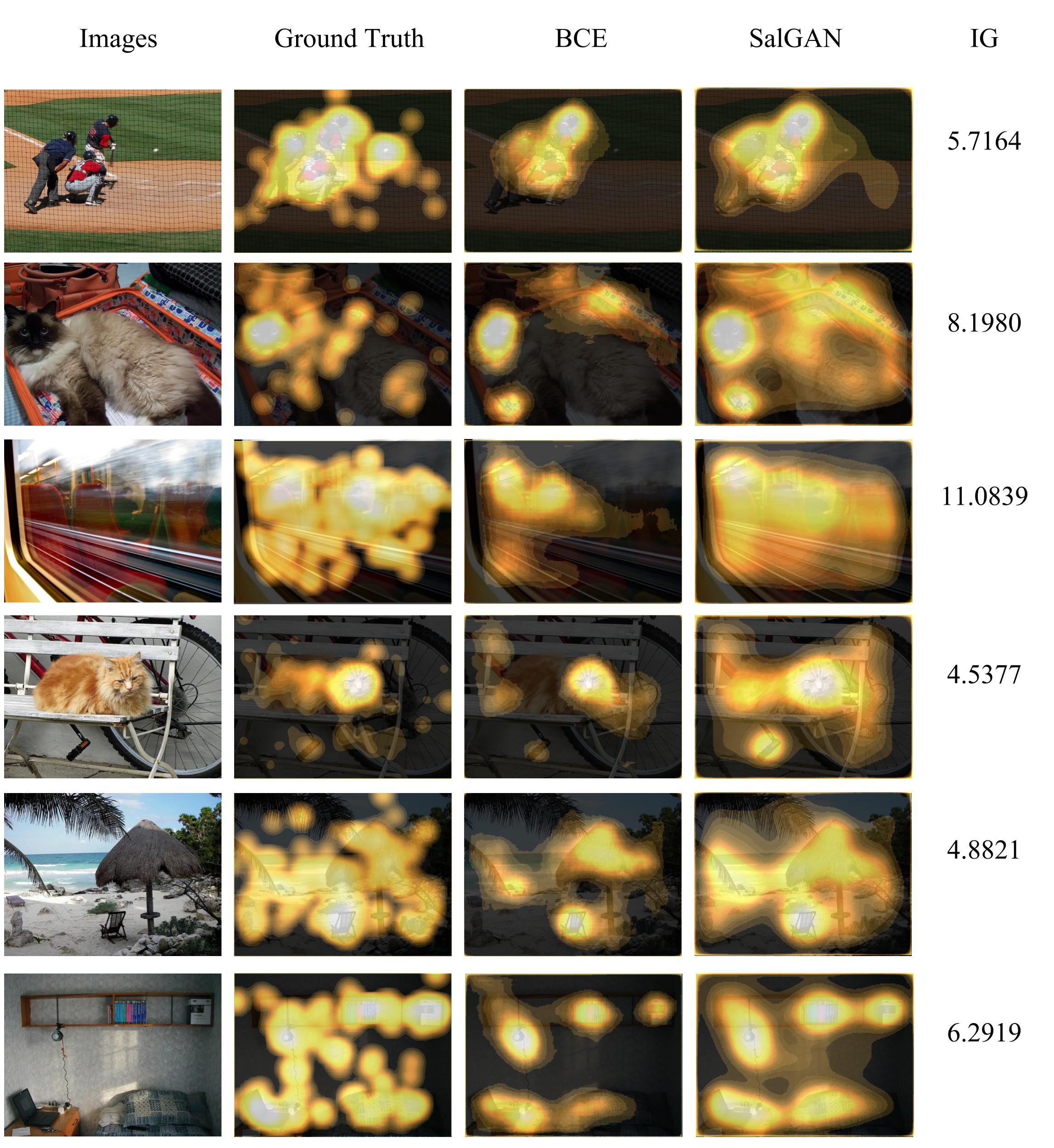
-
-
-
-## Datasets
-
-### Training
-As explained in our paper, our networks were trained on the training and validation data provided by [SALICON](http://salicon.net/).
-
-### Test
-Two different dataset were used for test:
-* Test partition of [SALICON](http://salicon.net/) dataset.
-* [MIT300](http://saliency.mit.edu/datasets.html).
-
-
-## Software frameworks
-
-Our paper presents two convolutional neural networks, one correspends to the Generator (Saliency Prediction Network) and the another is the Discriminator for the adversarial training. To compute saliency maps only the Generator is needed.
-
-### SalGAN on Lasagne
-
-SalGAN is implemented in [Lasagne](https://github.com/Lasagne/Lasagne), which at its time is developed over [Theano](http://deeplearning.net/software/theano/).
-```
-pip install -r https://github.com/imatge-upc/saliency-salgan-2017/blob/junting/requirements.txt
-```
-
-### Usage
-
-To train our model from scrath you need to run the following command:
-```
-THEANO_FLAGS=mode=FAST_RUN,device=gpu,floatX=float32,lib.cnmem=1,optimizer_including=cudnn python 02-train.py
-```
-In order to run the test script to predict saliency maps, you can run the following command after specifying the path to you images and the path to the output saliency maps:
-```
-THEANO_FLAGS=mode=FAST_RUN,device=gpu,floatX=float32,lib.cnmem=1,optimizer_including=cudnn python 03-predict.py
-```
-With the provided model weights you should obtain the follwing result:
-
-| ![Image Stimuli] | ![Saliency Map] |
-|:-:|:-:|
-
-[Image Stimuli]:https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/master/images/i112.jpg
-[Saliency Map]:https://raw.githubusercontent.com/imatge-upc/saliency-salgan-2017/master/saliency/i112.jpg
-
-Download the pretrained VGG-16 weights from: [vgg16.pkl](https://s3.amazonaws.com/lasagne/recipes/pretrained/imagenet/vgg16.pkl)
-
-
-## Acknowledgements
-
-We would like to especially thank Albert Gil Moreno and Josep Pujal from our technical support team at the Image Processing Group at the UPC.
-
-| ![AlbertGil-photo] | ![JosepPujal-photo] |
-|:-:|:-:|
-| [Albert Gil](AlbertGil-web) | [Josep Pujal](JosepPujal-web) |
-
-[AlbertGil-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/AlbertGil.jpg "Albert Gil"
-[JosepPujal-photo]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/authors/JosepPujal.jpg "Josep Pujal"
-
-[AlbertGil-web]: https://imatge.upc.edu/web/people/albert-gil-moreno
-[JosepPujal-web]: https://imatge.upc.edu/web/people/josep-pujal
-
-| | |
-|:--|:-:|
-| We gratefully acknowledge the support of [NVIDIA Corporation](http://www.nvidia.com/content/global/global.php) with the donation of the GeoForce GTX [Titan Z](http://www.nvidia.com/gtx-700-graphics-cards/gtx-titan-z/) and [Titan X](http://www.geforce.com/hardware/desktop-gpus/geforce-gtx-titan-x) used in this work. | ![logo-nvidia] |
-| The Image ProcessingGroup at the UPC is a [SGR14 Consolidated Research Group](https://imatge.upc.edu/web/projects/sgr14-image-and-video-processing-group) recognized and sponsored by the Catalan Government (Generalitat de Catalunya) through its [AGAUR](http://agaur.gencat.cat/en/inici/index.html) office. | ![logo-catalonia] |
-| This work has been developed in the framework of the projects [BigGraph TEC2013-43935-R](https://imatge.upc.edu/web/projects/biggraph-heterogeneous-information-and-graph-signal-processing-big-data-era-application) and [Malegra TEC2016-75976-R](https://imatge.upc.edu/web/projects/malegra-multimodal-signal-processing-and-machine-learning-graphs), funded by the Spanish Ministerio de Economía y Competitividad and the European Regional Development Fund (ERDF). | ![logo-spain] |
-| This publication has emanated from research conducted with the financial support of Science Foundation Ireland (SFI) under grant number SFI/12/RC/2289. | ![logo-ireland] |
-
-[logo-nvidia]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/nvidia.jpg "Logo of NVidia"
-[logo-catalonia]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/generalitat.jpg "Logo of Catalan government"
-[logo-spain]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/MEyC.png "Logo of Spanish government"
-[logo-ireland]: https://raw.githubusercontent.com/imatge-upc/saliency-2016-cvpr/master/logos/sfi.png "Logo of Science Foundation Ireland"
-
-## Contact
-
-If you have any general doubt about our work or code which may be of interest for other researchers, please use the [public issues section](https://github.com/imatge-upc/saliency-salgan-2017/issues) on this github repo. Alternatively, drop us an e-mail at .
-
-
-
-
diff --git a/authors/AlbertGil.jpg b/authors/AlbertGil.jpg
deleted file mode 100644
index b1fef00..0000000
Binary files a/authors/AlbertGil.jpg and /dev/null differ
diff --git a/authors/CristianCanton.jpg b/authors/CristianCanton.jpg
deleted file mode 100644
index 9fe40bd..0000000
Binary files a/authors/CristianCanton.jpg and /dev/null differ
diff --git a/authors/ElisaSayrol.jpg b/authors/ElisaSayrol.jpg
deleted file mode 100644
index a147fc5..0000000
Binary files a/authors/ElisaSayrol.jpg and /dev/null differ
diff --git a/authors/JordiTorres.jpg b/authors/JordiTorres.jpg
deleted file mode 100644
index fdcd552..0000000
Binary files a/authors/JordiTorres.jpg and /dev/null differ
diff --git a/authors/JosepPujal.jpg b/authors/JosepPujal.jpg
deleted file mode 100644
index e8c8cdb..0000000
Binary files a/authors/JosepPujal.jpg and /dev/null differ
diff --git a/authors/JuntingPan.jpg b/authors/JuntingPan.jpg
deleted file mode 100644
index ecb8c17..0000000
Binary files a/authors/JuntingPan.jpg and /dev/null differ
diff --git a/authors/Kevin160x160 2.jpg b/authors/Kevin160x160 2.jpg
deleted file mode 100644
index d97e5c4..0000000
Binary files a/authors/Kevin160x160 2.jpg and /dev/null differ
diff --git a/authors/KevinMcGuinness.jpg b/authors/KevinMcGuinness.jpg
deleted file mode 100644
index 2d2497e..0000000
Binary files a/authors/KevinMcGuinness.jpg and /dev/null differ
diff --git a/authors/KevinMcGuinness.png b/authors/KevinMcGuinness.png
deleted file mode 100644
index ec5f634..0000000
Binary files a/authors/KevinMcGuinness.png and /dev/null differ
diff --git a/authors/NoelOConnor.jpg b/authors/NoelOConnor.jpg
deleted file mode 100644
index b9d7274..0000000
Binary files a/authors/NoelOConnor.jpg and /dev/null differ
diff --git a/authors/NoelOConnor.png b/authors/NoelOConnor.png
deleted file mode 100644
index efa90ef..0000000
Binary files a/authors/NoelOConnor.png and /dev/null differ
diff --git a/authors/XavierGiro.jpg b/authors/XavierGiro.jpg
deleted file mode 100644
index 04e29dd..0000000
Binary files a/authors/XavierGiro.jpg and /dev/null differ
diff --git a/data/.gitignore b/data/.gitignore
new file mode 100644
index 0000000..94548af
--- /dev/null
+++ b/data/.gitignore
@@ -0,0 +1,3 @@
+*
+*/
+!.gitignore
diff --git a/figs/fullarchitecture.jpg b/figs/fullarchitecture.jpg
deleted file mode 100644
index ecc47a1..0000000
Binary files a/figs/fullarchitecture.jpg and /dev/null differ
diff --git a/figs/qualitative.jpg b/figs/qualitative.jpg
deleted file mode 100644
index f0dbcab..0000000
Binary files a/figs/qualitative.jpg and /dev/null differ
diff --git a/figs/thumbnail.jpg b/figs/thumbnail.jpg
deleted file mode 100644
index ca107a9..0000000
Binary files a/figs/thumbnail.jpg and /dev/null differ
diff --git a/figs/thumbnails.jpg b/figs/thumbnails.jpg
deleted file mode 100644
index 3132b3d..0000000
Binary files a/figs/thumbnails.jpg and /dev/null differ
diff --git a/images/i112.jpg b/images/i112.jpg
deleted file mode 100644
index 8bbddf2..0000000
Binary files a/images/i112.jpg and /dev/null differ
diff --git a/logos/MEyC.png b/logos/MEyC.png
deleted file mode 100644
index 2fede4d..0000000
Binary files a/logos/MEyC.png and /dev/null differ
diff --git a/logos/bsc320x86.jpg b/logos/bsc320x86.jpg
deleted file mode 100644
index bad6e6c..0000000
Binary files a/logos/bsc320x86.jpg and /dev/null differ
diff --git a/logos/cvpr2016.jpg b/logos/cvpr2016.jpg
deleted file mode 100644
index fd59e44..0000000
Binary files a/logos/cvpr2016.jpg and /dev/null differ
diff --git a/logos/dcu.png b/logos/dcu.png
deleted file mode 100644
index 8549d96..0000000
Binary files a/logos/dcu.png and /dev/null differ
diff --git a/logos/etsetb.png b/logos/etsetb.png
deleted file mode 100644
index 98176dc..0000000
Binary files a/logos/etsetb.png and /dev/null differ
diff --git a/logos/generalitat.jpg b/logos/generalitat.jpg
deleted file mode 100644
index 3dbb96e..0000000
Binary files a/logos/generalitat.jpg and /dev/null differ
diff --git a/logos/gpi.png b/logos/gpi.png
deleted file mode 100644
index 3b6429d..0000000
Binary files a/logos/gpi.png and /dev/null differ
diff --git a/logos/insight.jpg b/logos/insight.jpg
deleted file mode 100644
index aa2044c..0000000
Binary files a/logos/insight.jpg and /dev/null differ
diff --git a/logos/microsoft.jpg b/logos/microsoft.jpg
deleted file mode 100644
index 22951fc..0000000
Binary files a/logos/microsoft.jpg and /dev/null differ
diff --git a/logos/nvidia.jpg b/logos/nvidia.jpg
deleted file mode 100644
index 1540202..0000000
Binary files a/logos/nvidia.jpg and /dev/null differ
diff --git a/logos/sfi.png b/logos/sfi.png
deleted file mode 100644
index 27ec905..0000000
Binary files a/logos/sfi.png and /dev/null differ
diff --git a/logos/upc.jpg b/logos/upc.jpg
deleted file mode 100644
index 3c13a58..0000000
Binary files a/logos/upc.jpg and /dev/null differ
diff --git a/saliency/i112.jpg b/saliency/i112.jpg
deleted file mode 100644
index c82a7b9..0000000
Binary files a/saliency/i112.jpg and /dev/null differ
diff --git a/scripts/00-data_preparation.py b/scripts/00-data_preparation.py
new file mode 100644
index 0000000..fc844fe
--- /dev/null
+++ b/scripts/00-data_preparation.py
@@ -0,0 +1,58 @@
+import os
+import numpy as np
+from PIL import Image
+from PIL import ImageOps
+from scipy import misc
+import scipy.io
+from skimage import io
+import cv2
+import sys
+import cPickle as pickle
+import glob
+import random
+from tqdm import tqdm
+from eliaLib import dataRepresentation
+from constants import *
+from PIL import Image
+from PIL import ImageOps
+import pdb
+
+
+def augment_data():
+ listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages, '*'))]
+ listFilesTrain = [k for k in listImgFiles if 'train' in k]
+ listFilesVal = [k for k in listImgFiles if 'train' not in k]
+ for filenames in tqdm(listFilesTrain):
+ for angle in [90, 180, 270]:
+ src_im = Image.open(os.path.join(pathToImages,filenames+'.png'))
+ gt_im = Image.open(os.path.join(pathToMaps,filenames+'mask.png'))
+ rot_im = src_im.rotate(angle,expand=True)
+ rot_gt = gt_im.rotate(angle,expand=True)
+ rot_im.save(os.path.join(pathToImages,filenames+'_'+str(angle)+'.png'))
+ rot_gt.save(os.path.join(pathToMaps,filenames+'_'+str(angle)+'mask.png'))
+ vert_im = ImageOps.flip(src_im)
+ vert_gt = ImageOps.flip(gt_im)
+ horz_im = ImageOps.mirror(src_im)
+ horz_gt = ImageOps.mirror(gt_im)
+ vert_im.save(os.path.join(pathToImages,filenames+'_vert.png'))
+ vert_gt.save(os.path.join(pathToMaps,filenames+'_vertmask.png'))
+ horz_im.save(os.path.join(pathToImages,filenames+'_horz.png'))
+ horz_gt.save(os.path.join(pathToMaps,filenames+'_horzmask.png'))
+
+def split_data(fraction):
+ listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages, '*'))]
+ numSamples = len(listImgFiles)
+ train_ind = np.random.choice(np.arange(0,numSamples),(1,np.ceil(fraction*numSamples)),replace=False).squeeze()
+ val_ind = np.array(np.setdiff1d(np.arange(0,numSamples),train_ind)).squeeze()
+
+ for k in train_ind:
+ os.rename(os.path.join(pathToImages,listImgFiles[k] + '.png'),os.path.join(pathToImages,'train_'+listImgFiles[k]+'.png'))
+ os.rename(os.path.join(pathToMaps,listImgFiles[k] + 'mask.png'),os.path.join(pathToMaps,'train_'+listImgFiles[k]+'mask.png'))
+ for k in val_ind:
+ os.rename(os.path.join(pathToImages,listImgFiles[k] + '.png'),os.path.join(pathToImages,'val_'+listImgFiles[k]+'.png'))
+ os.rename(os.path.join(pathToMaps,listImgFiles[k] + 'mask.png'),os.path.join(pathToMaps,'val_'+listImgFiles[k]+'mask.png'))
+def main():
+# split_data(0.8)
+ augment_data()
+if __name__== "__main__":
+ main()
diff --git a/scripts/01-data_preprocessing.py b/scripts/01-data_preprocessing.py
index b82713d..825b627 100644
--- a/scripts/01-data_preprocessing.py
+++ b/scripts/01-data_preprocessing.py
@@ -14,46 +14,45 @@
from tqdm import tqdm
from eliaLib import dataRepresentation
from constants import *
-
+import pdb
img_size = INPUT_SIZE
salmap_size = INPUT_SIZE
# Resize train/validation files
-listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToMaps, '*'))]
-listTestImages = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages, '*test*'))]
-
-for currFile in tqdm(listImgFiles):
- tt = dataRepresentation.Target(os.path.join(pathToImages, currFile + '.jpg'),
- os.path.join(pathToMaps, currFile + '.mat'),
- os.path.join(pathToFixationMaps, currFile + '.mat'),
- dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
- dataRepresentation.LoadState.loaded, dataRepresentation.InputType.saliencyMapMatlab,
- dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty)
-
- # if tt.image.getImage().shape[:2] != (480, 640):
- # print 'Error:', currFile
-
- imageResized = cv2.cvtColor(cv2.resize(tt.image.getImage(), img_size, interpolation=cv2.INTER_AREA),
- cv2.COLOR_RGB2BGR)
- saliencyResized = cv2.resize(tt.saliency.getImage(), salmap_size, interpolation=cv2.INTER_AREA)
-
- cv2.imwrite(os.path.join(pathOutputImages, currFile + '.png'), imageResized)
- cv2.imwrite(os.path.join(pathOutputMaps, currFile + '.png'), saliencyResized)
+listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImagesNoAugment, '*'))]
+#listTestImages = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages, '*test*'))]
+#for currFile in tqdm(listImgFiles):
+# tt = dataRepresentation.Target(os.path.join(pathToImages, currFile + '.png'),
+# os.path.join(pathToMaps, currFile + 'mask.png'),
+# os.path.join(pathToFixationMaps, currFile + '.mat'),
+# dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
+# dataRepresentation.LoadState.loaded, dataRepresentation.InputType.imageGrayscale,
+# dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty)
+#
+# # if tt.image.getImage().shape[:2] != (480, 640):
+# # print 'Error:', currFile
+#
+# imageResized = cv2.cvtColor(cv2.resize(tt.image.getImage(), img_size, interpolation=cv2.INTER_AREA),
+# cv2.COLOR_RGB2BGR)
+# saliencyResized = cv2.resize(tt.saliency.getImage(), salmap_size, interpolation=cv2.INTER_AREA)
+#
+# cv2.imwrite(os.path.join(pathOutputImages, currFile + '.png'), imageResized)
+# cv2.imwrite(os.path.join(pathOutputMaps, currFile + 'mask.png'), saliencyResized)
# Resize test files
-for currFile in tqdm(listTestImages):
- tt = dataRepresentation.Target(os.path.join(pathToImages, currFile + '.jpg'),
- os.path.join(pathToMaps, currFile + '.mat'),
- os.path.join(pathToFixationMaps, currFile + '.mat'),
- dataRepresentation.LoadState.loaded,dataRepresentation.InputType.image,
- dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty,
- dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty)
-
- imageResized = cv2.cvtColor(cv2.resize(tt.image.getImage(), img_size, interpolation=cv2.INTER_AREA),
- cv2.COLOR_RGB2BGR)
- cv2.imwrite(os.path.join(pathOutputImages, currFile + '.png'), imageResized)
+#for currFile in tqdm(listTestImages):
+# tt = dataRepresentation.Target(os.path.join(pathToImages, currFile + '.jpg'),
+# os.path.join(pathToMaps, currFile + '.mat'),
+# os.path.join(pathToFixationMaps, currFile + '.mat'),
+# dataRepresentation.LoadState.loaded,dataRepresentation.InputType.image,
+# dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty,
+# dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty)
+#
+# imageResized = cv2.cvtColor(cv2.resize(tt.image.getImage(), img_size, interpolation=cv2.INTER_AREA),
+# cv2.COLOR_RGB2BGR)
+# cv2.imwrite(os.path.join(pathOutputImages, currFile + '.png'), imageResized)
# LOAD DATA
@@ -63,41 +62,43 @@
listFilesTrain = [k for k in listImgFiles if 'train' in k]
trainData = []
for currFile in tqdm(listFilesTrain):
- trainData.append(dataRepresentation.Target(os.path.join(pathOutputImages, currFile + '.png'),
- os.path.join(pathOutputMaps, currFile + '.png'),
+ trainData.append(dataRepresentation.Target(os.path.join(pathToImagesNoAugment, currFile + '.png'),
+ os.path.join(pathToMapsNoAugment, currFile + 'mask.png'),
os.path.join(pathToFixationMaps, currFile + '.mat'),
dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
dataRepresentation.LoadState.loaded, dataRepresentation.InputType.imageGrayscale,
- dataRepresentation.LoadState.loaded, dataRepresentation.InputType.fixationMapMatlab))
+ dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty))
-with open(os.path.join(pathToPickle, 'trainData.pickle'), 'wb') as f:
+with open(os.path.join(pathToPickle, 'trainDataNoAugment.pickle'), 'wb') as f:
pickle.dump(trainData, f)
# Validation
+listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages, '*'))]
listFilesValidation = [k for k in listImgFiles if 'val' in k]
validationData = []
for currFile in tqdm(listFilesValidation):
- validationData.append(dataRepresentation.Target(os.path.join(pathOutputImages, currFile + '.png'),
- os.path.join(pathOutputMaps, currFile + '.png'),
+ validationData.append(dataRepresentation.Target(os.path.join(pathToImages, currFile + '.png'),
+ os.path.join(pathToMaps, currFile + 'mask.png'),
os.path.join(pathToFixationMaps, currFile + '.mat'),
dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
dataRepresentation.LoadState.loaded, dataRepresentation.InputType.imageGrayscale,
- dataRepresentation.LoadState.loaded, dataRepresentation.InputType.fixationMapMatlab))
+ dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty))
with open(os.path.join(pathToPickle, 'validationData.pickle'), 'wb') as f:
pickle.dump(validationData, f)
# Test
-
+listFilesTest = [k for k in listImgFiles if 'test' in k]
testData = []
-
-for currFile in tqdm(listTestImages):
- testData.append(dataRepresentation.Target(os.path.join(pathOutputImages, currFile + '.png'),
- os.path.join(pathOutputMaps, currFile + '.png'),
- dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
- dataRepresentation.LoadState.unloaded,
- dataRepresentation.InputType.empty))
+for currFile in tqdm(listFilesTest):
+ testData.append(dataRepresentation.Target(os.path.join(pathToImages, currFile + '.png'),
+ os.path.join(pathToMaps, currFile + 'mask.png'),
+ os.path.join(pathToFixationMaps, currFile + '.mat'),
+ dataRepresentation.LoadState.loaded, dataRepresentation.InputType.image,
+ dataRepresentation.LoadState.loaded, dataRepresentation.InputType.imageGrayscale,
+ dataRepresentation.LoadState.unloaded, dataRepresentation.InputType.empty))
with open(os.path.join(pathToPickle, 'testData.pickle'), 'wb') as f:
pickle.dump(testData, f)
+
diff --git a/scripts/02-train.py b/scripts/02-train.py
index 6b98a30..f6a545a 100644
--- a/scripts/02-train.py
+++ b/scripts/02-train.py
@@ -1,7 +1,3 @@
-# Two mode of training available:
-# - BCE: CNN training, NOT Adversarial Training here. Only learns the generator network.
-# - SALGAN: Adversarial Training. Updates weights for both Generator and Discriminator.
-# The training used data previously processed using "01-data_preocessing.py"
import os
import numpy as np
import sys
@@ -11,103 +7,144 @@
import theano
import theano.tensor as T
import lasagne
-
+from sympy.utilities.iterables import cartes
from tqdm import tqdm
from constants import *
from models.model_salgan import ModelSALGAN
from models.model_bce import ModelBCE
from utils import *
-
+import pdb
+import matplotlib
+
+#####################################
+#To bypass X11 for matplotlib in tmux
+matplotlib.use('Agg')
+import matplotlib.pyplot as plt
+#####################################
flag = str(sys.argv[1])
-
-def bce_batch_iterator(model, train_data, validation_sample):
- num_epochs = 301
+def bce_batch_iterator(model, train_data, validation_data, validation_sample, epochs = 10, fig=False):
+ num_epochs = epochs+1
n_updates = 1
nr_batches_train = int(len(train_data) / model.batch_size)
+ train_loss_plt, train_acc_plt, val_loss_plt, val_acc_plt = [[] for i in range(4)]
for current_epoch in tqdm(range(num_epochs), ncols=20):
- e_cost = 0.
-
+ counter = 0
+ e_cost = 0.;tr_acc = 0.; tr_loss = 0.
random.shuffle(train_data)
-
for currChunk in chunks(train_data, model.batch_size):
-
if len(currChunk) != model.batch_size:
continue
-
- batch_input = np.asarray([x.image.data.astype(theano.config.floatX).transpose(2, 0, 1) for x in currChunk],
- dtype=theano.config.floatX)
-
- batch_output = np.asarray([y.saliency.data.astype(theano.config.floatX) / 255. for y in currChunk],
- dtype=theano.config.floatX)
+ batch_input = np.asarray([x.image.data.astype(theano.config.floatX).transpose(2, 0, 1) for x in currChunk],dtype=theano.config.floatX)
+ batch_output = np.asarray([y.saliency.data.astype(theano.config.floatX) / 255. for y in currChunk],dtype=theano.config.floatX)
batch_output = np.expand_dims(batch_output, axis=1)
-
- # train generator with one batch and discriminator with next batch
G_cost = model.G_trainFunction(batch_input, batch_output)
- e_cost += G_cost
+ if counter < 20:
+ tr_l, tr_jac = model.G_valFunction(batch_input,batch_output)
+ tr_loss += tr_l; tr_acc += tr_jac
+ counter += 1
+ e_cost += G_cost;
n_updates += 1
-
- e_cost /= nr_batches_train
-
- print 'Epoch:', current_epoch, ' train_loss->', e_cost
-
+ e_cost /= nr_batches_train; tr_acc /= counter; tr_loss /= counter
+ train_loss_plt.append(tr_loss);train_acc_plt.append(tr_acc)
+ print '\n train_loss->', e_cost
+ print ' train_accuracy(subset)->', tr_acc
+
+ v_cost, v_acc = bce_feedforward(model,validation_data,True)
+ val_loss_plt.append(v_cost);val_acc_plt.append(v_acc)
if current_epoch % 5 == 0:
+ if fig is True:
+ draw_figs(train_loss_plt, val_loss_plt, 'Train Loss', 'Val Loss')
+ draw_figs(train_acc_plt, val_acc_plt, 'Train Acc', 'Val Acc')
np.savez('./' + DIR_TO_SAVE + '/gen_modelWeights{:04d}.npz'.format(current_epoch),
*lasagne.layers.get_all_param_values(model.net['output']))
- predict(model=model, image_stimuli=validation_sample, num_epoch=current_epoch, path_output_maps=DIR_TO_SAVE)
-
+ predict(model=model, image_stimuli=validation_sample, num_epoch=current_epoch, path_output_maps=FIG_SAVE_DIR)
+ return v_acc
-def salgan_batch_iterator(model, train_data, validation_sample):
- num_epochs = 301
+def salgan_batch_iterator(model, train_data, validation_data,validation_sample,epochs = 20, fig=False):
+ num_epochs = epochs+1
nr_batches_train = int(len(train_data) / model.batch_size)
+ train_loss_plt, train_acc_plt, val_loss_plt, val_acc_plt = [[] for i in range(4)]
n_updates = 1
for current_epoch in tqdm(range(num_epochs), ncols=20):
-
- g_cost = 0.
- d_cost = 0.
- e_cost = 0.
-
+ g_cost = 0.; d_cost = 0.; e_cost = 0.
random.shuffle(train_data)
-
for currChunk in chunks(train_data, model.batch_size):
-
if len(currChunk) != model.batch_size:
continue
-
- batch_input = np.asarray([x.image.data.astype(theano.config.floatX).transpose(2, 0, 1) for x in currChunk],
- dtype=theano.config.floatX)
- batch_output = np.asarray([y.saliency.data.astype(theano.config.floatX) / 255. for y in currChunk],
- dtype=theano.config.floatX)
+ batch_input = np.asarray([x.image.data.astype(theano.config.floatX).transpose(2, 0, 1) for x in currChunk],dtype=theano.config.floatX)
+ batch_output = np.asarray([y.saliency.data.astype(theano.config.floatX) / 255. for y in currChunk],dtype=theano.config.floatX)
batch_output = np.expand_dims(batch_output, axis=1)
-
- # train generator with one batch and discriminator with next batch
if n_updates % 2 == 0:
G_obj, D_obj, G_cost = model.G_trainFunction(batch_input, batch_output)
- d_cost += D_obj
- g_cost += G_obj
- e_cost += G_cost
+ d_cost += D_obj; g_cost += G_obj; e_cost += G_cost
else:
G_obj, D_obj, G_cost = model.D_trainFunction(batch_input, batch_output)
- d_cost += D_obj
- g_cost += G_obj
- e_cost += G_cost
-
+ d_cost += D_obj; g_cost += G_obj; e_cost += G_cost
n_updates += 1
-
g_cost /= nr_batches_train
- d_cost /= nr_batches_train
- e_cost /= nr_batches_train
+ d_cost /= nr_batches_train
+ e_cost /= nr_batches_train
+ #Compute the Jaccard Index on the Validation
+ v_cost, v_acc = bce_feedforward(model,validation_data,True)
- # Save weights every 3 epoch
- if current_epoch % 3 == 0:
+ if current_epoch % 5 == 0:
np.savez('./' + DIR_TO_SAVE + '/gen_modelWeights{:04d}.npz'.format(current_epoch),
*lasagne.layers.get_all_param_values(model.net['output']))
np.savez('./' + DIR_TO_SAVE + '/disrim_modelWeights{:04d}.npz'.format(current_epoch),
*lasagne.layers.get_all_param_values(model.discriminator['fc5']))
- predict(model=model, image_stimuli=validation_sample, numEpoch=current_epoch, pathOutputMaps=DIR_TO_SAVE)
- print 'Epoch:', current_epoch, ' train_loss->', (g_cost, d_cost, e_cost)
-
+ predict(model=model, image_stimuli=validation_sample, num_epoch=current_epoch, path_output_maps=FIG_SAVE_DIR)
+ return v_acc
+
+def bce_feedforward(model, validation_data, bPrint=False):
+ nr_batches_val = int(len(validation_data) / model.batch_size)
+ v_cost = 0.
+ v_acc = 0.
+ for currChunk in chunks(validation_data, model.batch_size):
+ if len(currChunk) != model.batch_size:
+ continue
+ batch_input = np.asarray([x.image.data.astype(theano.config.floatX).transpose(2, 0, 1) for x in currChunk],dtype=theano.config.floatX)
+ batch_output = np.asarray([y.saliency.data.astype(theano.config.floatX) / 255. for y in currChunk],dtype=theano.config.floatX)
+ batch_output = np.expand_dims(batch_output, axis=1)
+ val_loss, val_accuracy = model.G_valFunction(batch_input,batch_output)
+ v_cost += val_loss
+ v_acc += val_accuracy
+ v_cost /= nr_batches_val
+ v_acc /= nr_batches_val
+ if bPrint is True:
+ print " validation_accuracy -->", v_acc
+ print " validation_loss -->", v_cost
+ print "-----------------------------------------"
+ return v_cost, v_acc
+
+def draw_figs(x,y,current_epoch,label1,label2):
+ fig1 = plt.figure(1)
+ plt.plot(range(current_epoch+1),x,color='red',linestyle='-',label=label1)
+ plt.plot(range(current_epoch+1),val_loss_plt,color='blue',linestyle='-',label=label2)
+ if label == 'Train Loss':
+ plt.title("Train and Val loss");plt.xlabel("Epochs");plt.ylabel("Loss")
+ plt.legend()
+ plt.savefig('./'+FIG_SAVE_DIR+'/train_val_loss_{:04d}.png'.format(current_epoch))
+ plt.close(fig1)
+ else:
+ plt.title("Train and Val Accuracy");plt.xlabel("Epochs");plt.ylabel("Acc")
+ plt.legend()
+ plt.savefig('./'+FIG_SAVE_DIR+'/train_val_acc_{:04d}.png'.format(current_epoch))
+ plt.close(fig1)
+def test():
+ """
+ Tests generator on the test set
+ :return:
+ """
+ # Load data
+ print 'Loading test data...'
+ with open(TEST_DATA_DIR, 'rb') as f:
+ test_data = pickle.load(f)
+ print '-->done!'
+ model = ModelSALGAN(INPUT_SIZE[0], INPUT_SIZE[1],9,0.01,1e-05,0.01,0.2)
+ load_weights(net=model.net['output'], path='weights/gen_', epochtoload=15)
+ bce_feedforward(model,test_data,bPrint=True)
def train():
"""
Train both generator and discriminator
@@ -115,39 +152,90 @@ def train():
"""
# Load data
print 'Loading training data...'
- with open('../saliency-2016-lsun/validationSample240x320.pkl', 'rb') as f:
- # with open(TRAIN_DATA_DIR, 'rb') as f:
+ with open(TRAIN_DATA_DIR, 'rb') as f:
train_data = pickle.load(f)
print '-->done!'
- print 'Loading validation data...'
- with open('../saliency-2016-lsun/validationSample240x320.pkl', 'rb') as f:
- # with open(VALIDATION_DATA_DIR, 'rb') as f:
+ print 'Loading test data...'
+ with open(TEST_DATA_DIR, 'rb') as f:
validation_data = pickle.load(f)
print '-->done!'
# Choose a random sample to monitor the training
num_random = random.choice(range(len(validation_data)))
validation_sample = validation_data[num_random]
- cv2.imwrite('./' + DIR_TO_SAVE + '/validationRandomSaliencyGT.png', validation_sample.saliency.data)
- cv2.imwrite('./' + DIR_TO_SAVE + '/validationRandomImage.png', cv2.cvtColor(validation_sample.image.data,
+ cv2.imwrite('./' + FIG_SAVE_DIR + '/validationRandomSaliencyGT.png', validation_sample.saliency.data)
+ cv2.imwrite('./' + FIG_SAVE_DIR + '/validationRandomImage.png', cv2.cvtColor(validation_sample.image.data,
cv2.COLOR_RGB2BGR))
-
# Create network
-
if flag == 'salgan':
- model = ModelSALGAN(INPUT_SIZE[0], INPUT_SIZE[1])
+ model = ModelSALGAN(INPUT_SIZE[0], INPUT_SIZE[1],9,0.01,1e-05,0.01,0.2)
# Load a pre-trained model
- # load_weights(net=model.net['output'], path="nss/gen_", epochtoload=15)
- # load_weights(net=model.discriminator['fc5'], path="test_dialted/disrim_", epochtoload=54)
- salgan_batch_iterator(model, train_data, validation_sample.image.data)
+ load_weights(net=model.net['output'], path="weights/gen_", epochtoload=15)
+ load_weights(net=model.discriminator['fc5'], path="weights/disrim_", epochtoload=15)
+ salgan_batch_iterator(model, train_data, validation_data,validation_sample.image.data,epochs=5)
elif flag == 'bce':
- model = ModelBCE(INPUT_SIZE[0], INPUT_SIZE[1])
+ model = ModelBCE(INPUT_SIZE[0], INPUT_SIZE[1],10,0.05,1e-5,0.99)
# Load a pre-trained model
# load_weights(net=model.net['output'], path='test/gen_', epochtoload=15)
- bce_batch_iterator(model, train_data, validation_sample.image.data)
+ bce_batch_iterator(model, train_data, validation_data,validation_sample.image.data,epochs=10)
else:
print "Invalid input argument."
+def cross_val():
+ # Load data
+ print 'Loading training data...'
+ with open(TRAIN_DATA_DIR_CROSS, 'rb') as f:
+ train_data = pickle.load(f)
+ print '-->done!'
+
+ print 'Loading validation data...'
+ with open(VAL_DATA_DIR, 'rb') as f:
+ validation_data = pickle.load(f)
+ print '-->done!'
+ num_random = random.choice(range(len(validation_data)))
+ validation_sample = validation_data[num_random]
+ if flag == 'bce':
+ lr_list = [0.1,0.01,0.001,0.05]
+ regterm_list = [1e-1,1e-2,1e-3,1e-4,1e-5]
+ momentum_list = [0.9,0.99]
+ lr,regterm,mom,acc = [[] for i in range(4)]
+ for config_list in list(cartes(lr_list,regterm_list,momentum_list)):
+ model = ModelBCE(INPUT_SIZE[0], INPUT_SIZE[1],16,config_list[0],config_list[1],config_list[2])
+ val_accuracy = bce_batch_iterator(model, train_data, validation_data,validation_sample.image.data,epochs=10)
+ lr.append(config_list[0])
+ regterm.append(config_list[1])
+ mom.append(config_list[2])
+ acc.append(val_accuracy)
+ for l,r,m,a in zip(lr,regterm,mom,acc):
+ print ("lr: {}, lambda: {}, momentum: {}, accuracy: {}").format(l,r,m,a)
+ print('------------------------------------------------------------------')
+
+ print('--------------------------------The Best--------------------------')
+ best_idx = np.argmax(acc)
+ print ("lr: {}, lambda: {}, momentum: {}, accuracy: {}").format(lr[best_idx],regterm[best_idx],mom[best_idx],acc[best_idx])
+ elif flag == 'salgan':
+ G_lr_list = [0.1,0.01,0.05]
+ regterm_list = [1e-1,1e-2,1e-3,1e-4,1e-5]
+ D_lr_list = [0.1,0.01,0.05]
+ alpha_list = [1/5., 1/10., 1/20.]
+ G_lr,regterm,D_lr,alpha,acc = [[] for i in range(5)]
+ for config_list in list(cartes(G_lr_list,regterm_list,D_lr_list,alpha_list)):
+ model = ModelSALGAN(INPUT_SIZE[0], INPUT_SIZE[1],9,config_list[0],config_list[1],config_list[2],config_list[3])
+ val_accuracy = salgan_batch_iterator(model, train_data, validation_data,validation_sample.image.data,epochs=10)
+ G_lr.append(config_list[0])
+ regterm.append(config_list[1])
+ D_lr.append(config_list[2])
+ alpha.append(config_list[3])
+ acc.append(val_accuracy)
+ for g_l,r,d_l,al,a in zip(G_lr,regterm,D_lr,alpha,acc):
+ print ("G_lr: {}, lambda: {}, D_lr: {}, alpha: {}, accuracy: {}").format(g_l,r,d_l,al,a)
+ print('------------------------------------------------------------------')
+
+ print('--------------------------------The Best--------------------------')
+ best_idx = np.argmax(acc)
+ print ("G_lr: {}, lambda: {}, D_lr: {}, alpha: {}, accuracy: {}").format(G_lr[best_idx],regterm[best_idx],D_lr[best_idx],alpha[best_idx],acc[best_idx])
+ else:
+ print("Please provide a correct argument")
if __name__ == "__main__":
train()
diff --git a/scripts/03-predict.py b/scripts/03-predict.py
index 0701fc6..13712c8 100644
--- a/scripts/03-predict.py
+++ b/scripts/03-predict.py
@@ -6,25 +6,26 @@
from utils import *
from constants import *
from models.model_bce import ModelBCE
-
+from models.model_salgan import ModelSALGAN
+import pdb
def test(path_to_images, path_output_maps, model_to_test=None):
- list_img_files = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(path_to_images, '*'))]
+ list_img_files = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(path_to_images, 'test_*.png'))]
# Load Data
- list_img_files.sort()
- for curr_file in tqdm(list_img_files, ncols=20):
- print os.path.join(path_to_images, curr_file + '.jpg')
- img = cv2.cvtColor(cv2.imread(os.path.join(path_to_images, curr_file + '.jpg'), cv2.IMREAD_COLOR), cv2.COLOR_BGR2RGB)
+ for curr_file in tqdm(list_img_files):
+# print os.path.join(path_to_images, curr_file + '.png')
+ img = cv2.cvtColor(cv2.imread(os.path.join(path_to_images, curr_file + '.png'), cv2.IMREAD_COLOR), cv2.COLOR_BGR2RGB)
predict(model=model_to_test, image_stimuli=img, name=curr_file, path_output_maps=path_output_maps)
def main():
# Create network
- model = ModelBCE(INPUT_SIZE[0], INPUT_SIZE[1], batch_size=8)
+ model = ModelBCE(INPUT_SIZE[0], INPUT_SIZE[1],10,0.05,1e-5,0.99)
+ #model = ModelSALGAN(INPUT_SIZE[0], INPUT_SIZE[1],9,0.01,1e-05,0.01,0.2)
# Here need to specify the epoch of model sanpshot
- load_weights(model.net['output'], path='gen_', epochtoload=90)
+ load_weights(model.net['output'], path="bce_weights/gen_", epochtoload=10)
# Here need to specify the path to images and output path
- test(path_to_images='../images/', path_output_maps='../saliency/', model_to_test=model)
+ test(path_to_images=pathToImages, path_output_maps=pathToResMaps, model_to_test=model)
if __name__ == "__main__":
main()
diff --git a/scripts/04-evaluate.py b/scripts/04-evaluate.py
new file mode 100644
index 0000000..aaa8ca9
--- /dev/null
+++ b/scripts/04-evaluate.py
@@ -0,0 +1,151 @@
+import os
+import numpy as np
+import cv2
+import sys
+import glob
+from tqdm import tqdm
+from constants import *
+from sklearn.metrics import jaccard_similarity_score
+from sklearn.metrics import confusion_matrix
+
+def evaluate():
+ listImgFiles = [k.split('/')[-1].split('.')[0] for k in glob.glob(os.path.join(pathToImages,'test*.png'))]
+ jaccard_score=0.
+ dice = 0.
+ spec = 0.
+ sens = 0.
+ acc = 0.
+ for currFile in tqdm(listImgFiles):
+# res = np.float32(cv2.imread(os.path.join(pathToResMaps,currFile+'.png'),cv2.IMREAD_GRAYSCALE))/255
+ res = np.load(os.path.join(pathToResMaps,currFile+'_gan.npy'))
+ gt = np.float32(cv2.imread(os.path.join(pathToMaps,currFile+'mask.png'),cv2.IMREAD_GRAYSCALE))/255
+
+ jaccard_score += jaccard_similarity_coefficient(gt,res)
+ dice += dice_coefficient(gt, res)
+ spec_tmp, sens_tmp, acc_tmp = specificity_sensitivity(gt, res)
+ spec += spec_tmp
+ sens += sens_tmp
+ acc += acc_tmp
+
+ dice /= len(listImgFiles)
+ jaccard_score /= len(listImgFiles)
+ spec /= len(listImgFiles)
+ sens /= len(listImgFiles)
+ acc /= len(listImgFiles)
+
+ print "Accuracy: ", acc
+ print "Sensitivity: ", sens
+ print "Specificity: ", spec
+ print "Jaccard: ", jaccard_score
+ print "Dice: ", dice
+
+def dice_coefficient(gt, res):
+
+ A = gt.flatten()
+ B = res.flatten()
+
+ A = np.array([1 if x > 0.5 else 0.0 for x in A])
+ B = np.array([1 if x > 0.5 else 0.0 for x in B])
+ dice = np.sum(B[A==1.0])*2.0 / (np.sum(B) + np.sum(A))
+ return dice
+def specificity_sensitivity(gt, res):
+ A = gt.flatten()
+ B = res.flatten()
+
+ A = np.array([1 if x > 0.5 else 0.0 for x in A])
+ B = np.array([1 if x > 0.5 else 0.0 for x in B])
+
+ tn, fp, fn, tp = np.float32(confusion_matrix(A, B).ravel())
+ specificity = tn/(fp + tn)
+ sensitivity = tp / (tp + fn)
+ accuracy = (tp+tn)/(tp+fp+fn+tn)
+
+ return specificity, sensitivity,accuracy
+
+def jaccard_similarity_coefficient(A, B, no_positives=1.0):
+ """Returns the jaccard index/similarity coefficient between A and B.
+
+ This should work for arrays of any dimensions.
+
+ J = len(intersection(A,B)) / len(union(A,B))
+
+ To extend to probabilistic input, to compute the intersection, use the min(A,B).
+ To compute the union, use max(A,B).
+
+ Assumes that a value of 1 indicates the positive values.
+ A value of 0 indicates the negative values.
+
+ If no positive values (1) in either A or B, then returns no_positives.
+ """
+ # Make sure the shapes are the same.
+ if not A.shape == B.shape:
+ raise ValueError("A and B must be the same shape")
+
+
+ # Make sure values are between 0 and 1.
+ if np.any( (A>1.) | (A<0) | (B>1.) | (B<0)):
+ raise ValueError("A and B must be between 0 and 1")
+
+ # Flatten to handle nd arrays.
+ A = A.flatten()
+ B = B.flatten()
+
+ intersect = np.minimum(A,B)
+ union = np.maximum(A, B)
+
+ # Special case if neither A or B have a 1 value.
+ if union.sum() == 0:
+ return no_positives
+
+ # Compute the Jaccard.
+ J = float(intersect.sum()) / union.sum()
+ return J
+
+
+def _jaccard_similarity_coefficient():
+ A = np.asarray([0,0,1])
+ B = np.asarray([0,1,1])
+ exp_out = 1. / 2
+ act_out = jaccard_similarity_coefficient(A,B)
+ assert act_out == exp_out, "Incorrect jaccard calculation"
+
+ A = np.asarray([0,1,1])
+ B = np.asarray([0,1,1])
+ act_out = jaccard_similarity_coefficient(A,B)
+ assert act_out == 1., "If same, then is 1."
+
+ A = np.asarray([0,1,1,0])
+ B = np.asarray([0,1,1])
+ try: excep = False; jaccard_similarity_coefficient(A,B) # This should throw an error.
+ except: excep = True
+ assert excep, "Error with different sized inputs."
+
+ A = np.asarray([0,1,1.1])
+ B = np.asarray([0,1,1])
+ try: excep = False; jaccard_similarity_coefficient(A,B) # This should throw an error.
+ except: excep = True
+ assert excep, "Values should be between 0 and 1."
+
+ A = np.asarray([1,0,1])
+ B = np.asarray([0,1,1])
+ A2 = np.asarray([A,A])
+ B2 = np.asarray([B,B])
+ act_out = jaccard_similarity_coefficient(A2,B2)
+ assert act_out == 1. / (3), "Incorrect 2D jaccard calculation."
+
+ # Fuzzy values.
+ A = np.asarray([0.5,0,1])
+ B = np.asarray([0.6,1,1])
+ exp_out = 1.5 / (0.6+2)
+ act_out = jaccard_similarity_coefficient(A,B)
+ assert act_out == exp_out, "Incorrect fuzzy jaccard calculation."
+
+#_jaccard_similarity_coefficient()
+
+def main():
+ evaluate()
+
+if __name__ == '__main__':
+ main()
+
+
diff --git a/scripts/constants.py b/scripts/constants.py
index fb1b11d..b5d0029 100644
--- a/scripts/constants.py
+++ b/scripts/constants.py
@@ -1,26 +1,33 @@
# Work space directory
-HOME_DIR = '/imatge/jpan/saliency-salgan-2017/'
+HOME_DIR = '/home/titan/Saeed/saliency-salgan-2017/'
# Path to SALICON raw data
-pathToImages = '/home/users/jpang/salicon_data/images'
-pathToMaps = '/home/users/jpang/salicon_data/saliency'
-pathToFixationMaps = '/home/users/jpang/salicon_data/fixation'
-
+pathToImages = '/home/titan/Saeed/saliency-salgan-2017/data/dermoFitImage'
+pathToMaps = '/home/titan/Saeed/saliency-salgan-2017/data/dermoFitMasks'
+pathToImagesNoAugment = '/home/titan/Saeed/saliency-salgan-2017/data/train_img_cross'
+pathToMapsNoAugment = '/home/titan/Saeed/saliency-salgan-2017/data/train_mask_cross'
+pathToResMaps = '/home/titan/Saeed/saliency-salgan-2017/data/dermoFitResults'
+pathToFixationMaps = ''
# Path to processed data
-pathOutputImages = '/home/users/jpang/lsun2016/data/salicon/images320x240'
-pathOutputMaps = '/home/users/jpang/lsun2016/data/salicon/saliency320x240'
-pathToPickle = '/home/users/jpang/scratch-local/salicon_data/320x240'
+pathOutputImages = '/home/titan/Saeed/saliency-salgan-2017/data/image320x240'
+pathOutputMaps = '/home/titan/Saeed/saliency-salgan-2017/data/mask320x240'
+pathToPickle = '/home/titan/Saeed/saliency-salgan-2017/data/pickle320x240'
# Path to pickles which contains processed data
-TRAIN_DATA_DIR = '/home/users/jpang/scratch-local/salicon_data/320x240/fix_trainData.pickle'
-VAL_DATA_DIR = '/home/users/jpang/scratch-local/salicon_data/320x240/fix_validationData.pickle'
-TEST_DATA_DIR = '/home/users/jpang/scratch-local/salicon_data/256x192/testData.pickle'
+TRAIN_DATA_DIR = '/home/titan/Saeed/saliency-salgan-2017/data/pickle320x240/trainData.pickle'
+TRAIN_DATA_DIR_CROSS = '/home/titan/Saeed/saliency-salgan-2017/data/pickle320x240/trainDataNoAugment.pickle'
+VAL_DATA_DIR = '/home/titan/Saeed/saliency-salgan-2017/data/pickle320x240/validationData.pickle'
+TEST_DATA_DIR = '/home/titan/Saeed/saliency-salgan-2017/data/pickle320x240/testData.pickle'
# Path to vgg16 pre-trained weights
-PATH_TO_VGG16_WEIGHTS = '/imatge/jpan/saliency-salgan-2017/vgg16.pkl'
+PATH_TO_VGG16_WEIGHTS = '/home/titan/Saeed/saliency-salgan-2017/models/vgg16.pkl'
# Input image and saliency map size
-INPUT_SIZE = (256, 192)
+INPUT_SIZE = (320,240)
# Directory to keep snapshots
-DIR_TO_SAVE = 'test'
+DIR_TO_SAVE = '../weights'
+FIG_SAVE_DIR = '../figs'
+
+#Path to test images
+pathToTestImages = '/home/titan/Saeed/saliency-salgan-2017/images'
diff --git a/scripts/constants.pyc b/scripts/constants.pyc
deleted file mode 100644
index 5447d92..0000000
Binary files a/scripts/constants.pyc and /dev/null differ
diff --git a/scripts/eliaLib/dataRepresentation.py b/scripts/eliaLib/dataRepresentation.py
index ca4d49d..4ffe874 100644
--- a/scripts/eliaLib/dataRepresentation.py
+++ b/scripts/eliaLib/dataRepresentation.py
@@ -120,4 +120,4 @@ def __init__(self, imagePath, saliencyPath,fixationPath,
fixationState=LoadState.unloaded, fixationType=InputType.fixationMapMatlab):
self.image = ImageContainer(imagePath, imageType, imageState)
self.saliency = ImageContainer(saliencyPath, saliencyType, saliencyState)
- self.fixation = ImageContainer(fixationPath, fixationType, fixationState)
\ No newline at end of file
+ self.fixation = ImageContainer(fixationPath, fixationType, fixationState)
diff --git a/scripts/models/__init__.pyc b/scripts/models/__init__.pyc
deleted file mode 100644
index b8d7773..0000000
Binary files a/scripts/models/__init__.pyc and /dev/null differ
diff --git a/scripts/models/discriminator.py b/scripts/models/discriminator.py
index 2c2b719..1f21598 100644
--- a/scripts/models/discriminator.py
+++ b/scripts/models/discriminator.py
@@ -2,7 +2,8 @@
from lasagne.layers import Pool2DLayer as PoolLayer
from lasagne.layers import DenseLayer, InputLayer
from lasagne.nonlinearities import tanh, sigmoid
-
+from lasagne.layers import batch_norm
+from lasagne.init import GlorotUniform
import nn
@@ -18,40 +19,40 @@ def build(input_height, input_width, concat_var):
net = {'input': InputLayer((None, 4, input_height, input_width), input_var=concat_var)}
print "Input: {}".format(net['input'].output_shape[1:])
- net['merge'] = ConvLayer(net['input'], 3, 1, pad=0, flip_filters=False)
+ net['merge'] = batch_norm(ConvLayer(net['input'], 3, 1, pad=0, W=GlorotUniform(gain="relu"),flip_filters=False))
print "merge: {}".format(net['merge'].output_shape[1:])
- net['conv1'] = ConvLayer(net['merge'], 32, 3, pad=1)
+ net['conv1'] = batch_norm(ConvLayer(net['merge'], 32, 3, pad=1,W=GlorotUniform(gain="relu")))
print "conv1: {}".format(net['conv1'].output_shape[1:])
net['pool1'] = PoolLayer(net['conv1'], 4)
print "pool1: {}".format(net['pool1'].output_shape[1:])
- net['conv2_1'] = ConvLayer(net['pool1'], 64, 3, pad=1)
+ net['conv2_1'] = batch_norm(ConvLayer(net['pool1'], 64, 3, pad=1,W=GlorotUniform(gain="relu")))
print "conv2_1: {}".format(net['conv2_1'].output_shape[1:])
- net['conv2_2'] = ConvLayer(net['conv2_1'], 64, 3, pad=1)
+ net['conv2_2'] = batch_norm(ConvLayer(net['conv2_1'], 64, 3, pad=1,W=GlorotUniform(gain="relu")))
print "conv2_2: {}".format(net['conv2_2'].output_shape[1:])
net['pool2'] = PoolLayer(net['conv2_2'], 2)
print "pool2: {}".format(net['pool2'].output_shape[1:])
- net['conv3_1'] = nn.weight_norm(ConvLayer(net['pool2'], 64, 3, pad=1))
+ net['conv3_1'] = batch_norm(ConvLayer(net['pool2'], 64, 3, pad=1,W=GlorotUniform(gain="relu")))
print "conv3_1: {}".format(net['conv3_1'].output_shape[1:])
- net['conv3_2'] = nn.weight_norm(ConvLayer(net['conv3_1'], 64, 3, pad=1))
+ net['conv3_2'] = batch_norm(ConvLayer(net['conv3_1'], 64, 3, pad=1,W=GlorotUniform(gain="relu")))
print "conv3_2: {}".format(net['conv3_2'].output_shape[1:])
net['pool3'] = PoolLayer(net['conv3_2'], 2)
print "pool3: {}".format(net['pool3'].output_shape[1:])
- net['fc4'] = DenseLayer(net['pool3'], num_units=100, nonlinearity=tanh)
+ net['fc4'] = batch_norm(DenseLayer(net['pool3'], num_units=100,W=GlorotUniform(gain="relu")))
print "fc4: {}".format(net['fc4'].output_shape[1:])
- net['fc5'] = DenseLayer(net['fc4'], num_units=2, nonlinearity=tanh)
+ net['fc5'] = batch_norm(DenseLayer(net['fc4'], num_units=2,W=GlorotUniform(gain="relu")))
print "fc5: {}".format(net['fc5'].output_shape[1:])
- net['prob'] = DenseLayer(net['fc5'], num_units=1, nonlinearity=sigmoid)
+ net['prob'] = batch_norm(DenseLayer(net['fc5'], num_units=1, W=GlorotUniform(gain=1.0),nonlinearity=sigmoid))
print "prob: {}".format(net['prob'].output_shape[1:])
return net
diff --git a/scripts/models/generator.py b/scripts/models/generator.py
index ed499f1..419cb17 100644
--- a/scripts/models/generator.py
+++ b/scripts/models/generator.py
@@ -5,6 +5,7 @@
import lasagne
import cPickle
import vgg16
+import unet
from constants import PATH_TO_VGG16_WEIGHTS
@@ -16,8 +17,9 @@ def set_pretrained_weights(net, path_to_model_weights=PATH_TO_VGG16_WEIGHTS):
def build_encoder(input_height, input_width, input_var):
- encoder = vgg16.build(input_height, input_width, input_var)
- set_pretrained_weights(encoder)
+ # encoder = vgg16.build(input_height, input_width, input_var)
+ encoder = unet.build(input_height, input_width, input_var)
+ #set_pretrained_weights(encoder)
return encoder
@@ -81,5 +83,5 @@ def build_decoder(net):
def build(input_height, input_width, input_var):
encoder = build_encoder(input_height, input_width, input_var)
- generator = build_decoder(encoder)
- return generator
+ #generator = build_decoder(encoder)
+ return encoder
diff --git a/scripts/models/generator.pyc b/scripts/models/generator.pyc
deleted file mode 100644
index 0d8b780..0000000
Binary files a/scripts/models/generator.pyc and /dev/null differ
diff --git a/scripts/models/layers.pyc b/scripts/models/layers.pyc
deleted file mode 100644
index 4a66591..0000000
Binary files a/scripts/models/layers.pyc and /dev/null differ
diff --git a/scripts/models/model.pyc b/scripts/models/model.pyc
deleted file mode 100644
index 3089d99..0000000
Binary files a/scripts/models/model.pyc and /dev/null differ
diff --git a/scripts/models/model_bce.py b/scripts/models/model_bce.py
index aec1f80..ea3e39c 100644
--- a/scripts/models/model_bce.py
+++ b/scripts/models/model_bce.py
@@ -9,27 +9,28 @@
class ModelBCE(Model):
- def __init__(self, w, h, batch_size=32, lr=0.001):
+ def __init__(self, w, h, batch_size,lr,regterm,momentum):
super(ModelBCE, self).__init__(w, h, batch_size)
self.net = generator.build(self.inputHeight, self.inputWidth, self.input_var)
output_layer_name = 'output'
- prediction = lasagne.layers.get_output(self.net[output_layer_name])
- test_prediction = lasagne.layers.get_output(self.net[output_layer_name], deterministic=True)
- self.predictFunction = theano.function([self.input_var], test_prediction)
-
- output_var_pooled = T.signal.pool.pool_2d(self.output_var, (4, 4), mode="average_exc_pad", ignore_border=True)
- prediction_pooled = T.signal.pool.pool_2d(prediction, (4, 4), mode="average_exc_pad", ignore_border=True)
-
- bce = lasagne.objectives.binary_crossentropy(prediction_pooled, output_var_pooled).mean()
+ prediction = lasagne.layers.get_output(self.net[output_layer_name],deterministic=False)
+ # Only for VGG 16 (Upsampling)
+ #prediction = T.nnet.abstract_conv.bilinear_upsampling(prediction,16)
+ #output_var_pooled = T.signal.pool.pool_2d(self.output_var, (16, 16), mode="average_exc_pad", ignore_border=True)
+ bce = lasagne.objectives.binary_crossentropy(prediction, self.output_var).mean() + regterm * lasagne.regularization.regularize_network_params(self.net[output_layer_name], lasagne.regularization.l2)
train_err = bce
-
- # parameters update and training
G_params = lasagne.layers.get_all_params(self.net[output_layer_name], trainable=True)
self.G_lr = theano.shared(np.array(lr, dtype=theano.config.floatX))
- G_updates = lasagne.updates.nesterov_momentum(train_err, G_params, learning_rate=self.G_lr, momentum=0.5)
-
- self.G_trainFunction = theano.function(inputs=[self.input_var, self.output_var], outputs=train_err, updates=G_updates,
- allow_input_downcast=True)
+ G_updates = lasagne.updates.momentum(train_err, G_params, learning_rate=self.G_lr,momentum=momentum)
+ self.G_trainFunction = theano.function(inputs=[self.input_var, self.output_var], outputs=train_err, updates=G_updates)
+
+ test_prediction = lasagne.layers.get_output(self.net[output_layer_name],deterministic=True)
+ # Only for VGG 16 (Upsampling)
+ #test_prediction = T.nnet.abstract_conv.bilinear_upsampling(test_prediction,16)
+ test_loss = lasagne.objectives.binary_crossentropy(test_prediction,self.output_var).mean()
+ test_acc = lasagne.objectives.binary_jaccard_index(test_prediction,self.output_var).mean()
+ self.G_valFunction = theano.function(inputs=[self.input_var, self.output_var],outputs=[test_loss,test_acc])
+ self.predictFunction = theano.function([self.input_var], test_prediction)
diff --git a/scripts/models/model_bce.pyc b/scripts/models/model_bce.pyc
deleted file mode 100644
index e004082..0000000
Binary files a/scripts/models/model_bce.pyc and /dev/null differ
diff --git a/scripts/models/model_salgan.py b/scripts/models/model_salgan.py
index 0b97c66..f594740 100644
--- a/scripts/models/model_salgan.py
+++ b/scripts/models/model_salgan.py
@@ -10,43 +10,27 @@
class ModelSALGAN(Model):
- def __init__(self, w, h, batch_size=32, G_lr=3e-4, D_lr=3e-4, alpha=1/20.):
+ def __init__(self, w, h, batch_size, G_lr, regterm, D_lr, alpha):
super(ModelSALGAN, self).__init__(w, h, batch_size)
# Build Generator
self.net = generator.build(self.inputHeight, self.inputWidth, self.input_var)
-
- # Build Discriminator
- self.discriminator = discriminator.build(self.inputHeight, self.inputWidth,
- T.concatenate([self.output_var, self.input_var], axis=1))
-
- # Set prediction function
+ self.discriminator = discriminator.build(self.inputHeight, self.inputWidth,T.concatenate([self.output_var, self.input_var], axis=1))
output_layer_name = 'output'
prediction = lasagne.layers.get_output(self.net[output_layer_name])
- test_prediction = lasagne.layers.get_output(self.net[output_layer_name], deterministic=True)
- self.predictFunction = theano.function([self.input_var], test_prediction)
- disc_lab = lasagne.layers.get_output(self.discriminator['prob'],
- T.concatenate([self.output_var, self.input_var], axis=1))
- disc_gen = lasagne.layers.get_output(self.discriminator['prob'],
- T.concatenate([prediction, self.input_var], axis=1))
+ disc_lab = lasagne.layers.get_output(self.discriminator['prob'],T.concatenate([self.output_var, self.input_var], axis=1))
+ disc_gen = lasagne.layers.get_output(self.discriminator['prob'],T.concatenate([prediction, self.input_var], axis=1))
- # Downscale the saliency maps
- output_var_pooled = T.signal.pool.pool_2d(self.output_var, (4, 4), mode="average_exc_pad", ignore_border=True)
- prediction_pooled = T.signal.pool.pool_2d(prediction, (4, 4), mode="average_exc_pad", ignore_border=True)
- train_err = lasagne.objectives.binary_crossentropy(prediction_pooled, output_var_pooled).mean()
- + 1e-4 * lasagne.regularization.regularize_network_params(self.net[output_layer_name], lasagne.regularization.l2)
+ train_err = lasagne.objectives.binary_crossentropy(prediction, self.output_var).mean() + regterm * lasagne.regularization.regularize_network_params(self.net[output_layer_name], lasagne.regularization.l2)
# Define loss function and input data
ones = T.ones(disc_lab.shape)
zeros = T.zeros(disc_lab.shape)
- D_obj = lasagne.objectives.binary_crossentropy(T.concatenate([disc_lab, disc_gen], axis=0),
- T.concatenate([ones, zeros], axis=0)).mean()
- + 1e-4 * lasagne.regularization.regularize_network_params(self.discriminator['prob'], lasagne.regularization.l2)
+ D_obj = lasagne.objectives.binary_crossentropy(T.concatenate([disc_lab, disc_gen], axis=0),T.concatenate([ones, zeros], axis=0)).mean() + regterm * lasagne.regularization.regularize_network_params(self.discriminator['prob'], lasagne.regularization.l2)
- G_obj_d = lasagne.objectives.binary_crossentropy(disc_gen, T.ones(disc_lab.shape)).mean()
- + 1e-4 * lasagne.regularization.regularize_network_params(self.net[output_layer_name], lasagne.regularization.l2)
+ G_obj_d = lasagne.objectives.binary_crossentropy(disc_gen, T.ones(disc_lab.shape)).mean() + regterm * lasagne.regularization.regularize_network_params(self.net[output_layer_name], lasagne.regularization.l2)
G_obj = G_obj_d + train_err * alpha
cost = [G_obj, D_obj, train_err]
@@ -54,14 +38,18 @@ def __init__(self, w, h, batch_size=32, G_lr=3e-4, D_lr=3e-4, alpha=1/20.):
# parameters update and training of Generator
G_params = lasagne.layers.get_all_params(self.net[output_layer_name], trainable=True)
self.G_lr = theano.shared(np.array(G_lr, dtype=theano.config.floatX))
- G_updates = lasagne.updates.adagrad(G_obj, G_params, learning_rate=self.G_lr)
- self.G_trainFunction = theano.function(inputs=[self.input_var, self.output_var], outputs=cost,
- updates=G_updates, allow_input_downcast=True)
+ G_updates = lasagne.updates.momentum(G_obj, G_params, learning_rate=self.G_lr,momentum=0.99)
+ self.G_trainFunction = theano.function(inputs=[self.input_var, self.output_var], outputs=cost,updates=G_updates, allow_input_downcast=True)
# parameters update and training of Discriminator
D_params = lasagne.layers.get_all_params(self.discriminator['prob'], trainable=True)
self.D_lr = theano.shared(np.array(D_lr, dtype=theano.config.floatX))
- D_updates = lasagne.updates.adagrad(D_obj, D_params, learning_rate=self.D_lr)
- self.D_trainFunction = theano.function([self.input_var, self.output_var], cost, updates=D_updates,
- allow_input_downcast=True)
+ D_updates = lasagne.updates.momentum(D_obj, D_params, learning_rate=self.D_lr,momentum=0.99)
+ self.D_trainFunction = theano.function([self.input_var, self.output_var], cost, updates=D_updates,allow_input_downcast=True)
+
+ test_prediction = lasagne.layers.get_output(self.net[output_layer_name], deterministic=True)
+ test_loss = lasagne.objectives.binary_crossentropy(test_prediction,self.output_var).mean()
+ test_acc = lasagne.objectives.binary_jaccard_index(test_prediction,self.output_var).mean()
+ self.G_valFunction = theano.function(inputs=[self.input_var, self.output_var],outputs=[test_loss,test_acc])
+ self.predictFunction = theano.function([self.input_var], test_prediction)
diff --git a/scripts/models/unet.py b/scripts/models/unet.py
new file mode 100644
index 0000000..3cc1e69
--- /dev/null
+++ b/scripts/models/unet.py
@@ -0,0 +1,108 @@
+from collections import OrderedDict
+from lasagne.layers import (InputLayer, ConcatLayer, Pool2DLayer, ReshapeLayer, DimshuffleLayer, NonlinearityLayer,
+ DropoutLayer, Deconv2DLayer, batch_norm)
+try:
+ from lasagne.layers.dnn import Conv2DDNNLayer as ConvLayer
+except ImportError:
+ from lasagne.layers import Conv2DLayer as ConvLayer
+import lasagne
+from lasagne.init import HeNormal
+from lasagne.init import GlorotNormal
+from layers import RGBtoBGRLayer
+from lasagne.nonlinearities import sigmoid
+
+def build(inputHeight, inputWidth, input_var,do_dropout=False):
+ #net = OrderedDict()
+ net = {'input': InputLayer((None, 3, inputHeight, inputWidth), input_var=input_var)}
+ #net['input'] = InputLayer((None, 3, inputHeight, inputWidth), input_var=input_var)
+ print "Input: {}".format(net['input'].output_shape[1:])
+
+ net['bgr'] = RGBtoBGRLayer(net['input'])
+
+ net['contr_1_1'] = batch_norm(ConvLayer(net['bgr'], 64, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr1_1: {}".format(net['contr_1_1'].output_shape[1:])
+ net['contr_1_2'] = batch_norm(ConvLayer(net['contr_1_1'],64,3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr1_2: {}".format(net['contr_1_2'].output_shape[1:])
+ net['pool1'] = Pool2DLayer(net['contr_1_2'], 2)
+ print"pool1: {}".format(net['pool1'].output_shape[1:])
+
+
+ net['contr_2_1'] = batch_norm(ConvLayer(net['pool1'], 128, 3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr2_1: {}".format(net['contr_2_1'].output_shape[1:])
+ net['contr_2_2'] = batch_norm(ConvLayer(net['contr_2_1'], 128, 3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr2_2: {}".format(net['contr_2_2'].output_shape[1:])
+ net['pool2'] = Pool2DLayer(net['contr_2_2'], 2)
+ print "pool2: {}".format(net['pool2'].output_shape[1:])
+
+
+ net['contr_3_1'] = batch_norm(ConvLayer(net['pool2'],256, 3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr3_1: {}".format(net['contr_3_1'].output_shape[1:])
+ net['contr_3_2'] = batch_norm(ConvLayer(net['contr_3_1'], 256, 3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr3_2: {}".format(net['contr_3_2'].output_shape[1:])
+ net['pool3'] = Pool2DLayer(net['contr_3_2'], 2)
+ print "pool3: {}".format(net['pool3'].output_shape[1:])
+
+ net['contr_4_1'] = batch_norm(ConvLayer(net['pool3'], 512, 3, pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr4_1: {}".format(net['contr_4_1'].output_shape[1:])
+ net['contr_4_2'] = batch_norm(ConvLayer(net['contr_4_1'],512, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "convtr4_2: {}".format(net['contr_4_2'].output_shape[1:])
+ l = net['pool4'] = Pool2DLayer(net['contr_4_2'], 2)
+ print "pool4: {}".format(net['pool4'].output_shape[1:])
+ # the paper does not really describe where and how dropout is added. Feel free to try more options
+ if do_dropout:
+ l = DropoutLayer(l, p=0.4)
+
+ net['encode_1'] = batch_norm(ConvLayer(l,1024, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "encode_1: {}".format(net['encode_1'].output_shape[1:])
+ net['encode_2'] = batch_norm(ConvLayer(net['encode_1'], 1024, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "encode_2: {}".format(net['encode_2'].output_shape[1:])
+ net['upscale1'] = batch_norm(Deconv2DLayer(net['encode_2'],1024, 2, 2, crop="valid", W=GlorotNormal(gain="relu")))
+ print "upscale1: {}".format(net['upscale1'].output_shape[1:])
+
+ net['concat1'] = ConcatLayer([net['upscale1'], net['contr_4_2']], cropping=(None, None, "center", "center"))
+ print "concat1: {}".format(net['concat1'].output_shape[1:])
+ net['expand_1_1'] = batch_norm(ConvLayer(net['concat1'], 512, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "expand_1_1: {}".format(net['expand_1_1'].output_shape[1:])
+ net['expand_1_2'] = batch_norm(ConvLayer(net['expand_1_1'],512, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "expand_1_2: {}".format(net['expand_1_2'].output_shape[1:])
+ net['upscale2'] = batch_norm(Deconv2DLayer(net['expand_1_2'], 512, 2, 2, crop="valid", W=GlorotNormal(gain="relu")))
+ print "upscale2: {}".format(net['upscale2'].output_shape[1:])
+
+ net['concat2'] = ConcatLayer([net['upscale2'], net['contr_3_2']], cropping=(None, None, "center", "center"))
+ print "concat2: {}".format(net['concat2'].output_shape[1:])
+ net['expand_2_1'] = batch_norm(ConvLayer(net['concat2'], 256, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "expand_2_1: {}".format(net['expand_2_1'].output_shape[1:])
+ net['expand_2_2'] = batch_norm(ConvLayer(net['expand_2_1'], 256, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "expand_2_2: {}".format(net['expand_2_2'].output_shape[1:])
+ net['upscale3'] = batch_norm(Deconv2DLayer(net['expand_2_2'],256, 2, 2, crop="valid",W=GlorotNormal(gain="relu")))
+ print "upscale3: {}".format(net['upscale3'].output_shape[1:])
+
+ net['concat3'] = ConcatLayer([net['upscale3'], net['contr_2_2']], cropping=(None, None, "center", "center"))
+ print "concat3: {}".format(net['concat3'].output_shape[1:])
+ net['expand_3_1'] = batch_norm(ConvLayer(net['concat3'], 128, 3,pad='same',W=GlorotNormal(gain="relu")))
+ print "expand_3_1: {}".format(net['expand_3_1'].output_shape[1:])
+ net['expand_3_2'] = batch_norm(ConvLayer(net['expand_3_1'],128, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "expand_3_2: {}".format(net['expand_3_2'].output_shape[1:])
+ net['upscale4'] = batch_norm(Deconv2DLayer(net['expand_3_2'], 128, 2, 2, crop="valid", W=GlorotNormal(gain="relu")))
+ print "upscale4: {}".format(net['upscale4'].output_shape[1:])
+
+ net['concat4'] = ConcatLayer([net['upscale4'], net['contr_1_2']], cropping=(None, None, "center", "center"))
+ print "concat4: {}".format(net['concat4'].output_shape[1:])
+ net['expand_4_1'] = batch_norm(ConvLayer(net['concat4'], 64, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "expand_4_1: {}".format(net['expand_4_1'].output_shape[1:])
+ net['expand_4_2'] = batch_norm(ConvLayer(net['expand_4_1'],64, 3,pad='same', W=GlorotNormal(gain="relu")))
+ print "expand_4_2: {}".format(net['expand_4_2'].output_shape[1:])
+
+ net['output'] = ConvLayer(net['expand_4_2'],1, 1, nonlinearity=sigmoid)
+ print "output: {}".format(net['output'].output_shape[1:])
+# net['dimshuffle'] = DimshuffleLayer(net['output_segmentation'], (1, 0, 2, 3))
+# print "dimshuffle: {}".format(net['dimshuffle'].output_shape[1:])
+# net['reshapeSeg'] = ReshapeLayer(net['dimshuffle'], (2, -1))
+# print "reshapeSeg: {}".format(net['reshapeSeg'].output_shape[1:])
+# net['dimshuffle2'] = DimshuffleLayer(net['reshapeSeg'], (1, 0))
+# print "dimshuffle2: {}".format(net['dimshuffle2'].output_shape[1:])
+# net['output_flattened'] = NonlinearityLayer(net['dimshuffle2'], nonlinearity=lasagne.nonlinearities.softmax)
+# print "output_flattened: {}".format(net['output_flattened'].output_shape[1:])
+
+ return net
+
diff --git a/scripts/models/vgg16.py b/scripts/models/vgg16.py
index 8f2a6c4..8ec6cc0 100644
--- a/scripts/models/vgg16.py
+++ b/scripts/models/vgg16.py
@@ -7,6 +7,8 @@
from lasagne.layers import Pool2DLayer as PoolLayer
from lasagne.layers import InputLayer
from layers import RGBtoBGRLayer
+from lasagne.layers import batch_norm
+from lasagne.nonlinearities import sigmoid
def build(inputHeight, inputWidth, input_var):
@@ -23,82 +25,61 @@ def build(inputHeight, inputWidth, input_var):
net['bgr'] = RGBtoBGRLayer(net['input'])
- net['conv1_1'] = ConvLayer(net['bgr'], 64, 3, pad=1, flip_filters=False)
- net['conv1_1'].add_param(net['conv1_1'].W, net['conv1_1'].W.get_value().shape, trainable=False)
- net['conv1_1'].add_param(net['conv1_1'].b, net['conv1_1'].b.get_value().shape, trainable=False)
+ net['conv1_1'] = batch_norm(ConvLayer(net['bgr'], 64, 3, pad=1, flip_filters=False))
print "conv1_1: {}".format(net['conv1_1'].output_shape[1:])
- net['conv1_2'] = ConvLayer(net['conv1_1'], 64, 3, pad=1, flip_filters=False)
- net['conv1_2'].add_param(net['conv1_2'].W, net['conv1_2'].W.get_value().shape, trainable=False)
- net['conv1_2'].add_param(net['conv1_2'].b, net['conv1_2'].b.get_value().shape, trainable=False)
+ net['conv1_2'] = batch_norm(ConvLayer(net['conv1_1'], 64, 3, pad=1, flip_filters=False))
print "conv1_2: {}".format(net['conv1_2'].output_shape[1:])
net['pool1'] = PoolLayer(net['conv1_2'], 2)
print "pool1: {}".format(net['pool1'].output_shape[1:])
- net['conv2_1'] = ConvLayer(net['pool1'], 128, 3, pad=1, flip_filters=False)
- net['conv2_1'].add_param(net['conv2_1'].W, net['conv2_1'].W.get_value().shape, trainable=False)
- net['conv2_1'].add_param(net['conv2_1'].b, net['conv2_1'].b.get_value().shape, trainable=False)
+ net['conv2_1'] = batch_norm(ConvLayer(net['pool1'], 128, 3, pad=1, flip_filters=False))
print "conv2_1: {}".format(net['conv2_1'].output_shape[1:])
- net['conv2_2'] = ConvLayer(net['conv2_1'], 128, 3, pad=1, flip_filters=False)
- net['conv2_2'].add_param(net['conv2_2'].W, net['conv2_2'].W.get_value().shape, trainable=False)
- net['conv2_2'].add_param(net['conv2_2'].b, net['conv2_2'].b.get_value().shape, trainable=False)
+ net['conv2_2'] = batch_norm(ConvLayer(net['conv2_1'], 128, 3, pad=1, flip_filters=False))
print "conv2_2: {}".format(net['conv2_2'].output_shape[1:])
net['pool2'] = PoolLayer(net['conv2_2'], 2)
print "pool2: {}".format(net['pool2'].output_shape[1:])
- net['conv3_1'] = ConvLayer(net['pool2'], 256, 3, pad=1, flip_filters=False)
- net['conv3_1'].add_param(net['conv3_1'].W, net['conv3_1'].W.get_value().shape, trainable=False)
- net['conv3_1'].add_param(net['conv3_1'].b, net['conv3_1'].b.get_value().shape, trainable=False)
+ net['conv3_1'] = batch_norm(ConvLayer(net['pool2'], 256, 3, pad=1, flip_filters=False))
print "conv3_1: {}".format(net['conv3_1'].output_shape[1:])
- net['conv3_2'] = ConvLayer(net['conv3_1'], 256, 3, pad=1, flip_filters=False)
- net['conv3_2'].add_param(net['conv3_2'].W, net['conv3_2'].W.get_value().shape, trainable=False)
- net['conv3_2'].add_param(net['conv3_2'].b, net['conv3_2'].b.get_value().shape, trainable=False)
+ net['conv3_2'] = batch_norm(ConvLayer(net['conv3_1'], 256, 3, pad=1, flip_filters=False))
print "conv3_2: {}".format(net['conv3_2'].output_shape[1:])
- net['conv3_3'] = ConvLayer(net['conv3_2'], 256, 3, pad=1, flip_filters=False)
- net['conv3_3'].add_param(net['conv3_3'].W, net['conv3_3'].W.get_value().shape, trainable=False)
- net['conv3_3'].add_param(net['conv3_3'].b, net['conv3_3'].b.get_value().shape, trainable=False)
+ net['conv3_3'] = batch_norm(ConvLayer(net['conv3_2'], 256, 3, pad=1, flip_filters=False))
print "conv3_3: {}".format(net['conv3_3'].output_shape[1:])
net['pool3'] = PoolLayer(net['conv3_3'], 2)
print "pool3: {}".format(net['pool3'].output_shape[1:])
- net['conv4_1'] = ConvLayer(net['pool3'], 512, 3, pad=1, flip_filters=False)
- net['conv4_1'].add_param(net['conv4_1'].W, net['conv4_1'].W.get_value().shape)
- net['conv4_1'].add_param(net['conv4_1'].b, net['conv4_1'].b.get_value().shape)
+ net['conv4_1'] = batch_norm(ConvLayer(net['pool3'], 512, 3, pad=1, flip_filters=False))
print "conv4_1: {}".format(net['conv4_1'].output_shape[1:])
- net['conv4_2'] = ConvLayer(net['conv4_1'], 512, 3, pad=1, flip_filters=False)
- net['conv4_2'].add_param(net['conv4_2'].W, net['conv4_2'].W.get_value().shape)
- net['conv4_2'].add_param(net['conv4_2'].b, net['conv4_2'].b.get_value().shape)
+ net['conv4_2'] = batch_norm(ConvLayer(net['conv4_1'], 512, 3, pad=1, flip_filters=False))
print "conv4_2: {}".format(net['conv4_2'].output_shape[1:])
- net['conv4_3'] = ConvLayer(net['conv4_2'], 512, 3, pad=1, flip_filters=False)
- net['conv4_3'].add_param(net['conv4_3'].W, net['conv4_3'].W.get_value().shape)
- net['conv4_3'].add_param(net['conv4_3'].b, net['conv4_3'].b.get_value().shape)
+ net['conv4_3'] = batch_norm(ConvLayer(net['conv4_2'], 512, 3, pad=1, flip_filters=False))
print "conv4_3: {}".format(net['conv4_3'].output_shape[1:])
net['pool4'] = PoolLayer(net['conv4_3'], 2)
print "pool4: {}".format(net['pool4'].output_shape[1:])
- net['conv5_1'] = ConvLayer(net['pool4'], 512, 3, pad=1, flip_filters=False)
- net['conv5_1'].add_param(net['conv5_1'].W, net['conv5_1'].W.get_value().shape)
- net['conv5_1'].add_param(net['conv5_1'].b, net['conv5_1'].b.get_value().shape)
+ net['conv5_1'] = batch_norm(ConvLayer(net['pool4'], 512, 3, pad=1, flip_filters=False))
print "conv5_1: {}".format(net['conv5_1'].output_shape[1:])
- net['conv5_2'] = ConvLayer(net['conv5_1'], 512, 3, pad=1, flip_filters=False)
- net['conv5_2'].add_param(net['conv5_2'].W, net['conv5_2'].W.get_value().shape)
- net['conv5_2'].add_param(net['conv5_2'].b, net['conv5_2'].b.get_value().shape)
+ net['conv5_2'] = batch_norm(ConvLayer(net['conv5_1'], 512, 3, pad=1, flip_filters=False))
print "conv5_2: {}".format(net['conv5_2'].output_shape[1:])
- net['conv5_3'] = ConvLayer(net['conv5_2'], 512, 3, pad=1, flip_filters=False)
- net['conv5_3'].add_param(net['conv5_3'].W, net['conv5_3'].W.get_value().shape)
- net['conv5_3'].add_param(net['conv5_3'].b, net['conv5_3'].b.get_value().shape)
+ net['conv5_3'] = batch_norm(ConvLayer(net['conv5_2'], 512, 3, pad=1, flip_filters=False))
print "conv5_3: {}".format(net['conv5_3'].output_shape[1:])
+# net['pool5'] = PoolLayer(net['conv5_3'], 2)
+# print "pool5: {}".format(net['pool5'].output_shape[1:])
+
+ net['output'] = batch_norm(ConvLayer(net['conv5_3'], 1, 1, pad=0,nonlinearity=sigmoid))
+ print "output: {}".format(net['output'].output_shape[1:])
return net
diff --git a/scripts/models/vgg16.pyc b/scripts/models/vgg16.pyc
deleted file mode 100644
index dd33cb9..0000000
Binary files a/scripts/models/vgg16.pyc and /dev/null differ
diff --git a/scripts/utils.py b/scripts/utils.py
index 27ae88e..3441bad 100644
--- a/scripts/utils.py
+++ b/scripts/utils.py
@@ -4,7 +4,7 @@
import theano
import lasagne
from constants import HOME_DIR
-
+import pdb
def chunks(l, n):
"""Yield successive n-sized chunks from l."""
@@ -42,13 +42,15 @@ def predict(model, image_stimuli, num_epoch=None, name=None, path_output_maps=No
# resize back to original size
saliency_map = cv2.resize(saliency_map, size, interpolation=cv2.INTER_CUBIC)
# blur
- saliency_map = cv2.GaussianBlur(saliency_map, (blur_size, blur_size), 0)
+ #saliency_map = cv2.GaussianBlur(saliency_map, (blur_size, blur_size), 0)
# clip again
saliency_map = np.clip(saliency_map, 0, 255)
+
if name is None:
# When we use for testing, there is no file name provided.
cv2.imwrite('./' + path_output_maps + '/validationRandomSaliencyPred_{:04d}.png'.format(num_epoch), saliency_map)
else:
- cv2.imwrite(os.path.join(path_output_maps, name + '.jpg'), saliency_map)
+ cv2.imwrite(os.path.join(path_output_maps, name + '_unet.png'), saliency_map)
+ np.save(os.path.join(path_output_maps, name+'_unet'), result)
diff --git a/scripts/utils.pyc b/scripts/utils.pyc
deleted file mode 100644
index 295ca7d..0000000
Binary files a/scripts/utils.pyc and /dev/null differ