This part of the tutorial describes how to run a big data machine learning job on Google Cloud.
We will run the kMeans clustering on genomics variant data as in the 5.1_BigData_Genomics-Clustering
except that here will use the entire chromosome 22 file from the 1000 Genomes Project data set publicly hosted at Google Cloud.
To run this example you will need to apply for a Google Cloud account as well as upgrade to the 'non-trial' version. However since you receive the initial credit of ~$300 you can still run this example (any possibly other computations) for free.
In this example we use the australia-souteast1 region but the example can be run in any region since the 1000 Genomes data are available globally.
Google Cloud offers the service Dataproc for running Apache Spark jobs, which we will use to run our clustering example.
For Dataproc we need to convert our jupyter notebook into a command line python script by extracting the relevant code, adding command line argument handling and creation of the initial SparkSession. The final python script is available at: python/genomic_clustering.py
In order to make it available to `Dataproc' we need to upload it to Google Storage, together with the 'init' script from google-cloud/init-actions/install-pandas, which will install python pandas on each cluster node. Additional we also need to create the location for the output data produced by our script.
Create a Google storage bucket in your region of choice with the following structure and files inside:
<your-bucket>/
gc/
init-actions/
install-pandas
output/
python/
genomic_clustering.py
Your files will be available for Dataproc at the urls starting with gs://<your-bucket> for example is your is spark-test the output location url would be gs://spark-test/gc/output
Later in this tutorial whenever you see gs://spark-tutorial/...) replace it with gs://<your-bucket>/....
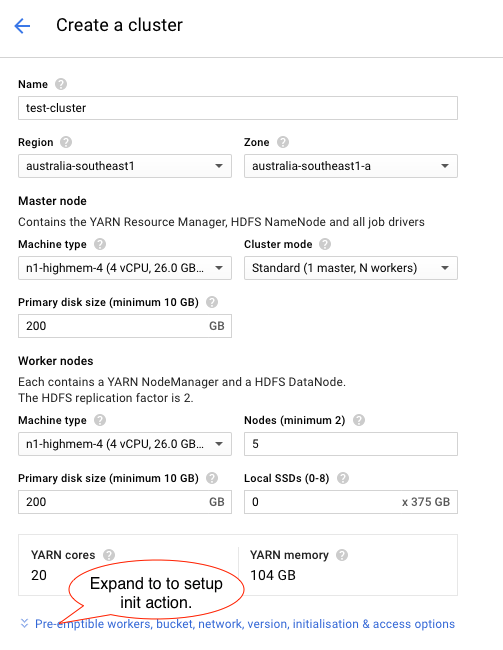
We will create a Dataproc cluster named test-cluster based on n1-himem-4 instances (4CPU cores) with one master node and 5 worker nodes (the type and number of instances are chosen to fit into the default quota of 24CPUs).
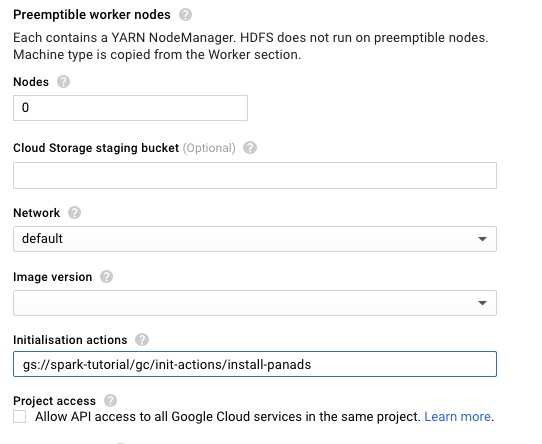
We will also setup the initialisation action for the cluster to install python pandas package in each node.
Using the Dataproc UI create the cluster with the following settings:
Make sure you also set the Initialisation action in the expandable section as shown below:
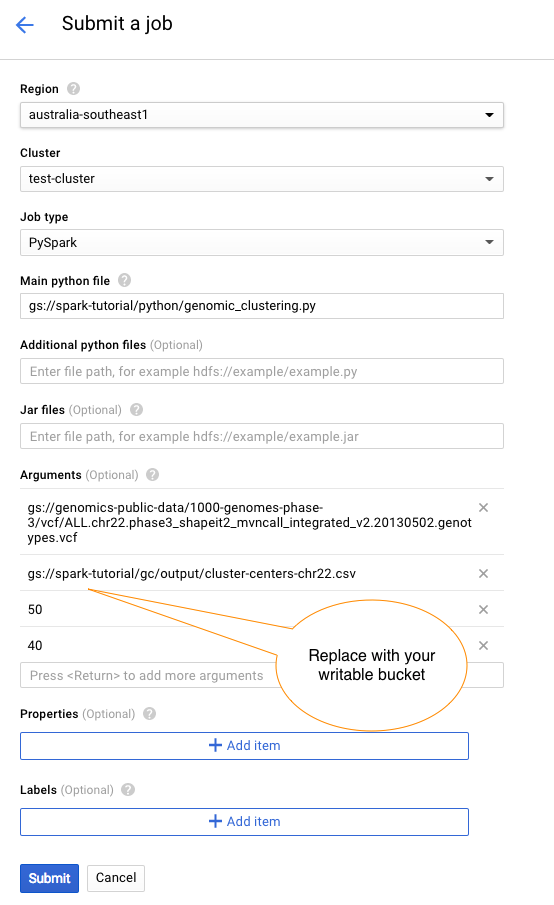
Once the cluster is up and and running we can submit a 'PySpark' job to run our python genomics clustering script.
Using the Dataproc UI submit a job with the following settings:
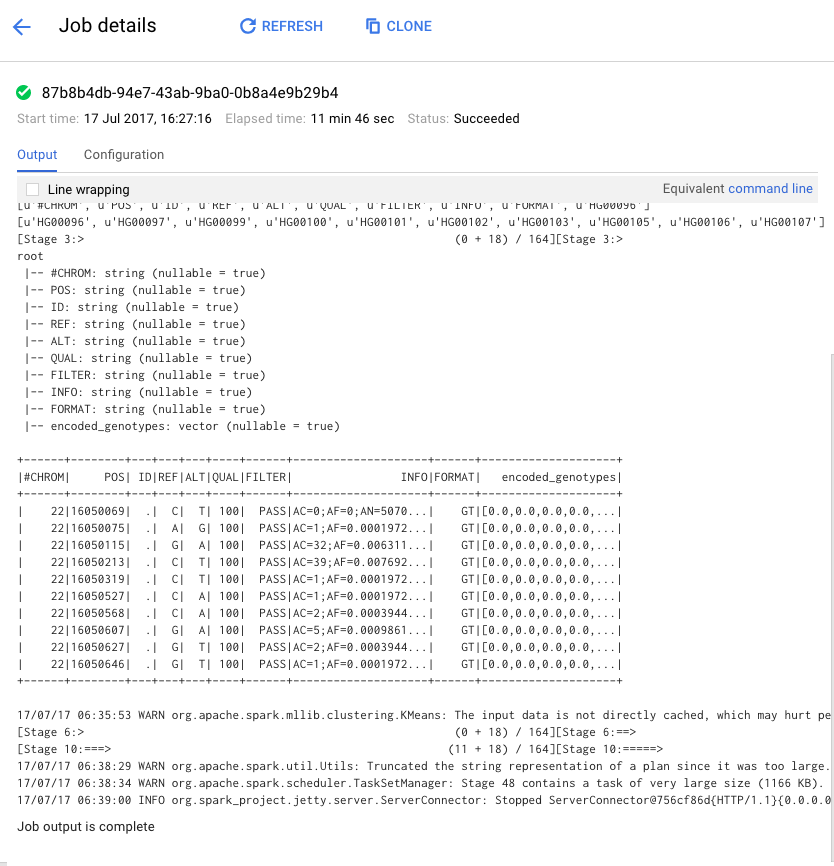
You can track progress of the job at the console. It should take about 10 minutes to complete.
The final results should be similar to the following:
Now you can download the results from Google Storage at: <your-bucket>/gc/output/cluster-centers-chr22.csv, and analyse them with 5.2_BigData_Genomics_Visualise.
Note:
Please remember to 'Delete' the cluster after you have finished running your jobs. If you keep it active you will be charged for the cloud usage even if you do not run any jobs!