-
Notifications
You must be signed in to change notification settings - Fork 3
Description
When processing some of my data, I encountered an issue with the computation of the snr_single. The issue occurs when the WM segmentation contains negative values.
Description
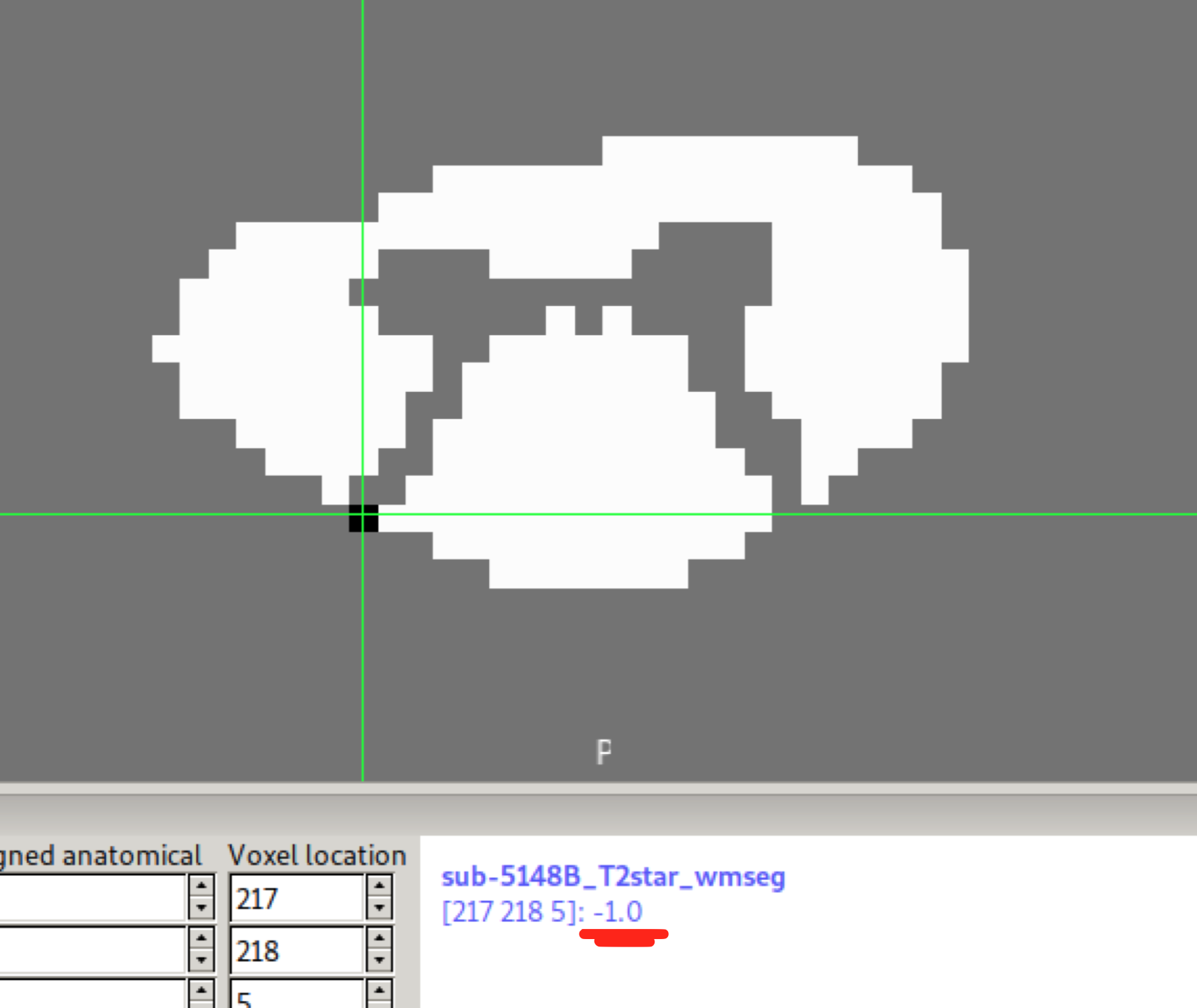
If the GM segmentation exceeds the SC segmentation:
The WM segmentation created by the sct_maths command:
Lines 125 to 126 in f8fd2b8
| # Generate white matter segmentation | |
| sct_maths -i ${file_1_seg}${ext} -sub ${file_1_gmseg}${ext} -o ${file_1}_wmseg${ext} |
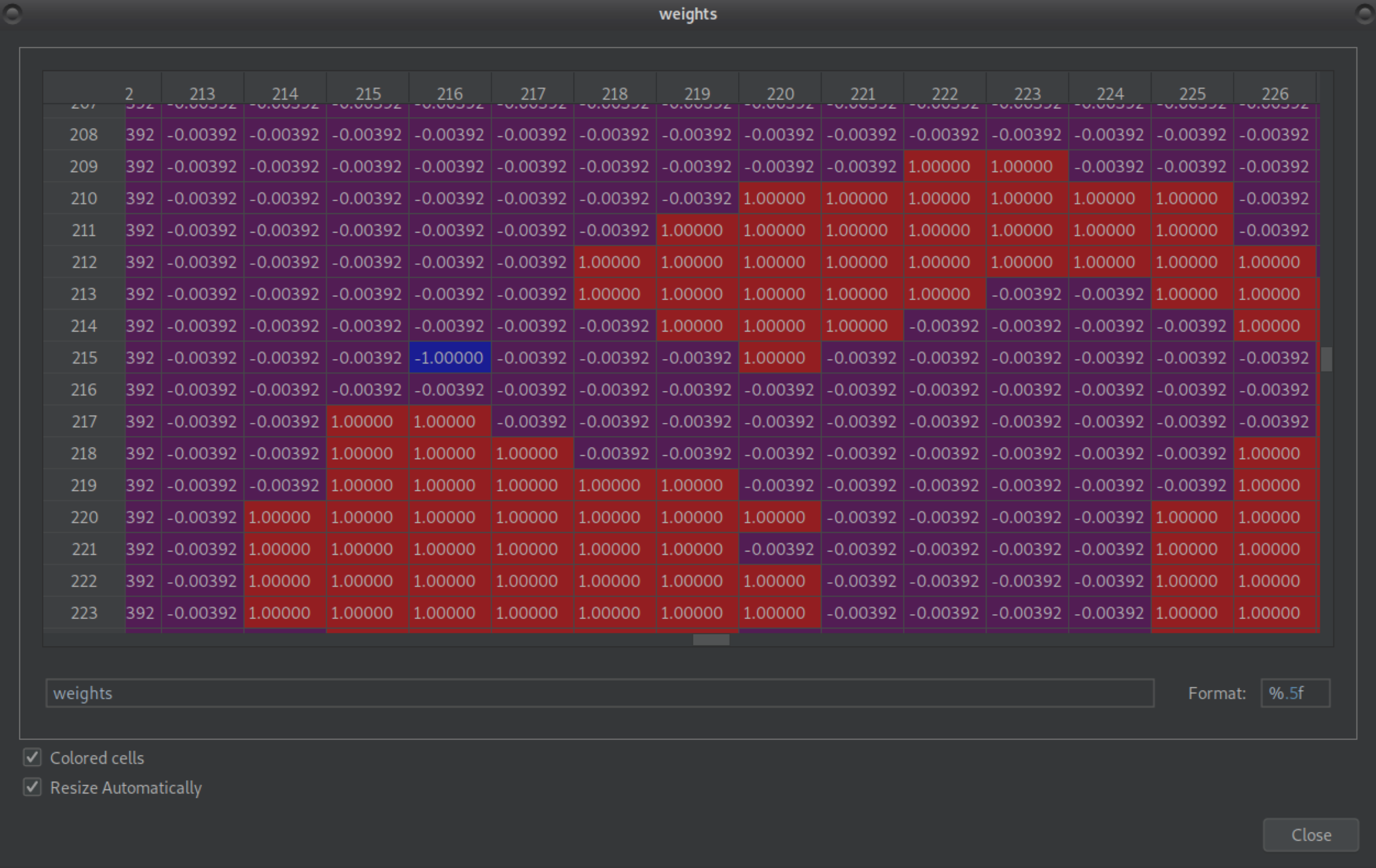
includes -1 value(s):
and also all voxels which were zero now have small negative values close to zero:
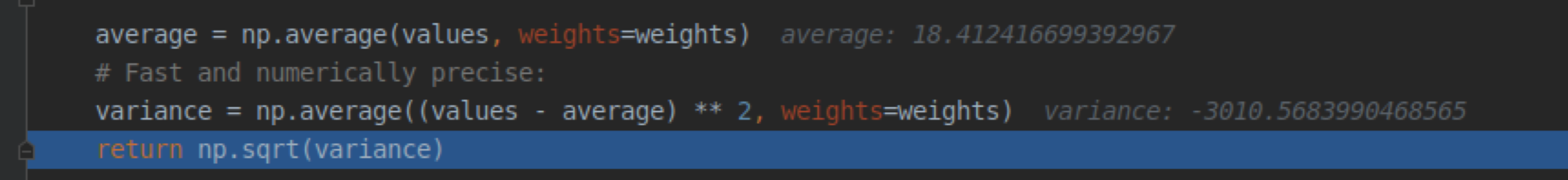
This results in a negative variance in the compute_cnr.py script:
gm-challenge/gmchallenge/compute_cnr.py
Lines 93 to 94 in f8fd2b8
| variance = np.average((values - average) ** 2, weights=weights) | |
| return np.sqrt(variance) |
for example:
which in turn causes that return np.sqrt(variance) returns NaN and computation of the noise_single_slice and snr_single_slice results also in NaN:
gm-challenge/gmchallenge/compute_cnr.py
Lines 112 to 114 in f8fd2b8
| noise_single_slice = \ | |
| [weighted_std(data1[..., iz], weights=mask[..., iz]) for iz in range(nz) if np.any(mask[..., iz])] | |
| snr_single_slice = [m / s for m, s in zip(mean_in_roi, noise_single_slice)] |
Workaround
The possible workaround which fixes the issue for me is the thresholding of the WM segmentation, for instance, like this:
sct_maths -i ${file_1}_wmseg${ext} -thr 0 -o ${file_1}_wmseg${ext}
SCT version
$ sct_check_dependencies -short
--
Spinal Cord Toolbox (git-HEAD-45b1c1759f991332697b4120160303e963f7a3c7)
sct_check_dependencies -short
--
SCT info:
- version: git-HEAD-45b1c1759f991332697b4120160303e963f7a3c7
- path: /usr/local/lib/sct_latest
OS: linux (Linux-5.10.0-0.bpo.9-amd64-x86_64-with-debian-10.12)
CPU cores: Available: 16, Used by ITK functions: 16
RAM: Total: 128744MB, Used: 15793MB, Available: 111367MB