-
Notifications
You must be signed in to change notification settings - Fork 41
Description
This is a simple HTML form to make use of the GBIF name parser. The parser is written in Java and based on regular expressions to dissect name strings into its components. It does only keep name parts required to reconstruct a full three-parted name with an optional subgenus, but ignores additional infraspecific parts such as the subspecies given for varieties. Please see our API documentation for details.
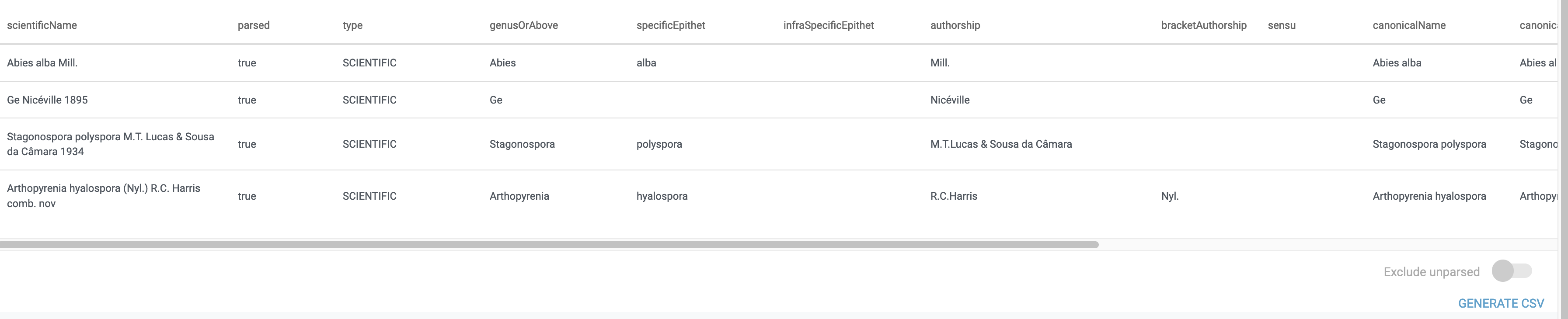
It takes data like this (imagine as single cells in a column):
Abies alba Mill.
Ge Nicéville 1895
Stagonospora polyspora M.T. Lucas & Sousa da Câmara 1934
Arthopyrenia hyalospora (Nyl.) R.C. Harris comb. nov
And can parse it into several columns:
After asking the #sp7-engineering channel internally:
Aimee: Yes, the specify network/broker uses. that, as well as the BISON project
Grant: Do you think it is something the Specify WorkBench could integrate to attempt to resolve unparsed taxonomy data?
Aimee: Yes, it’s reliable and you can send one or a list of names at once
Grant: Thank you 😄– I think it’s worth considering as nearly all of our conversions force users to parse that information before it can be imported and adds complexity to the process of getting data in Specify
Aimee: Good idea!