SeuSelect is a tool to make cell selections from a scRNA-seq experiment via a Seurat object. It is a Shiny based application.
library(devtools)
install_github("GhislainFievet/SeurSelect")and load the library
library(seurselect)You need a Seurat object, let's say pbmc.
You can get a Seurat object example:
download.file("https://www.dropbox.com/s/0xbzyfpr3eruapu/pbmc.RData?raw=1","pbmc.RData")
load("pbmc.RData")
pbmcTo run the app execute:
my.selections = SeurSelect(pmbc)On application exit, cell selections are stored in my.selections variable.
In this example from the demo we show how to select and export cells from the Seurat VlnPlot.
- Click create a selection
To create a new selection click on the create a selection button.
- Select the VlnPlot tab
You can make a selection from 4 different Seurat plots:
- Select the gene and the metadata to display on the plot, and click OK
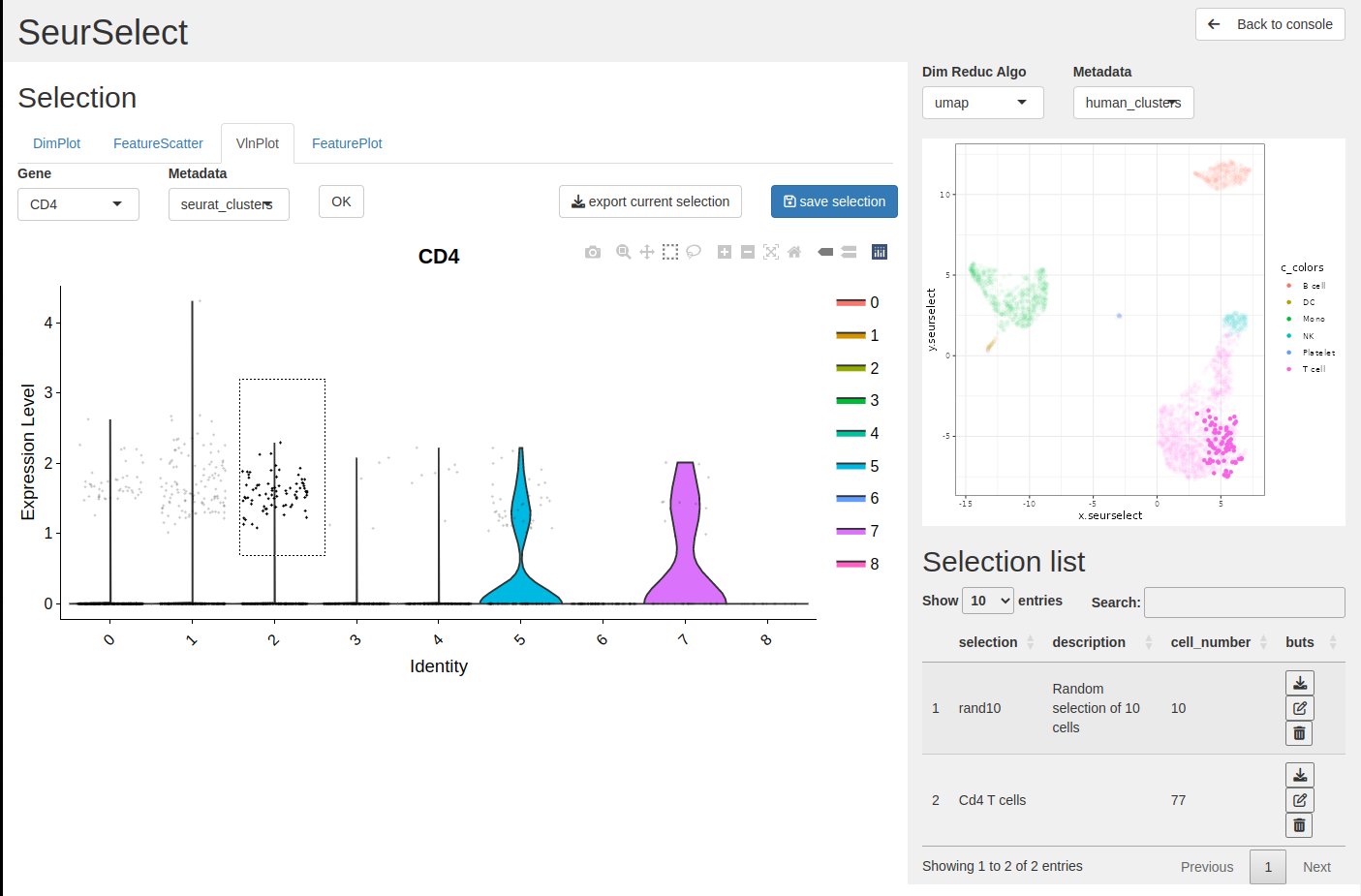
We want to select the CD4+ from the T cell group so we choose gene = CD4 and metadata = human_clusters.
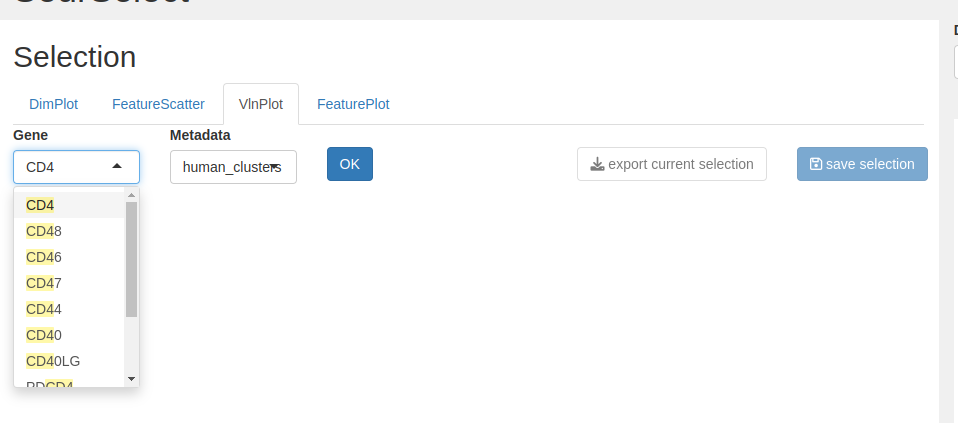
- Select from the interactive plot
With the select tool you can select the cells you want, here the CD4+ from the T cells type. You can see the selected cells on the right plot.
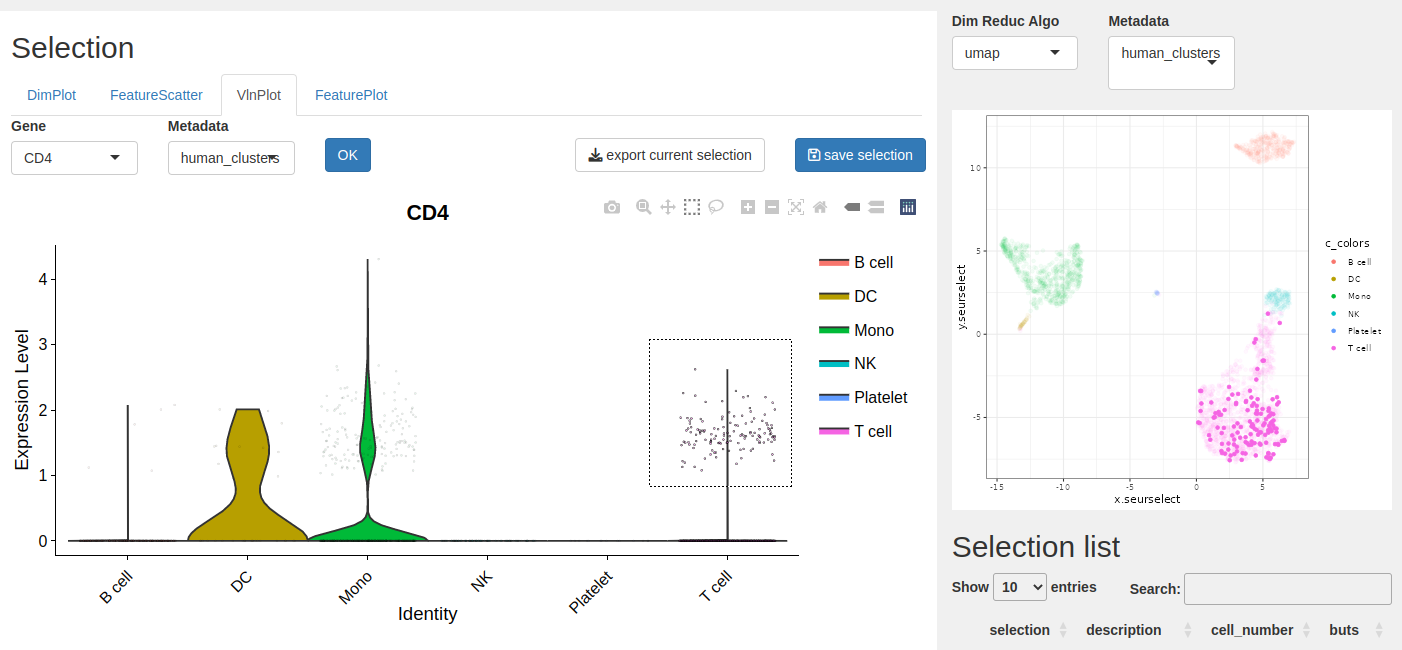
- Click save selection to store the list on the table.
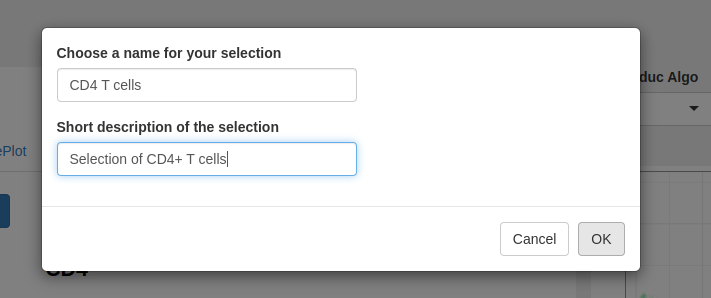
Provide a name, a description (optional) and click OK to add the selection on the list.
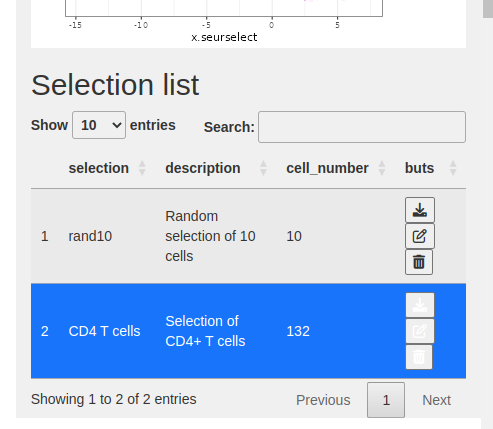
- The selection is added to the selection table.
- Click on Back to console button to exit the application.
On application exit, cells selections are stored in the variablemy.selections