ℹ️ Beta Version Available: A beta version with compatibility for Google Colab and newer versions of torch and scvi-tools is available on the
beta-colabbranch. Install it withpip install celldisect==0.2.0b1.
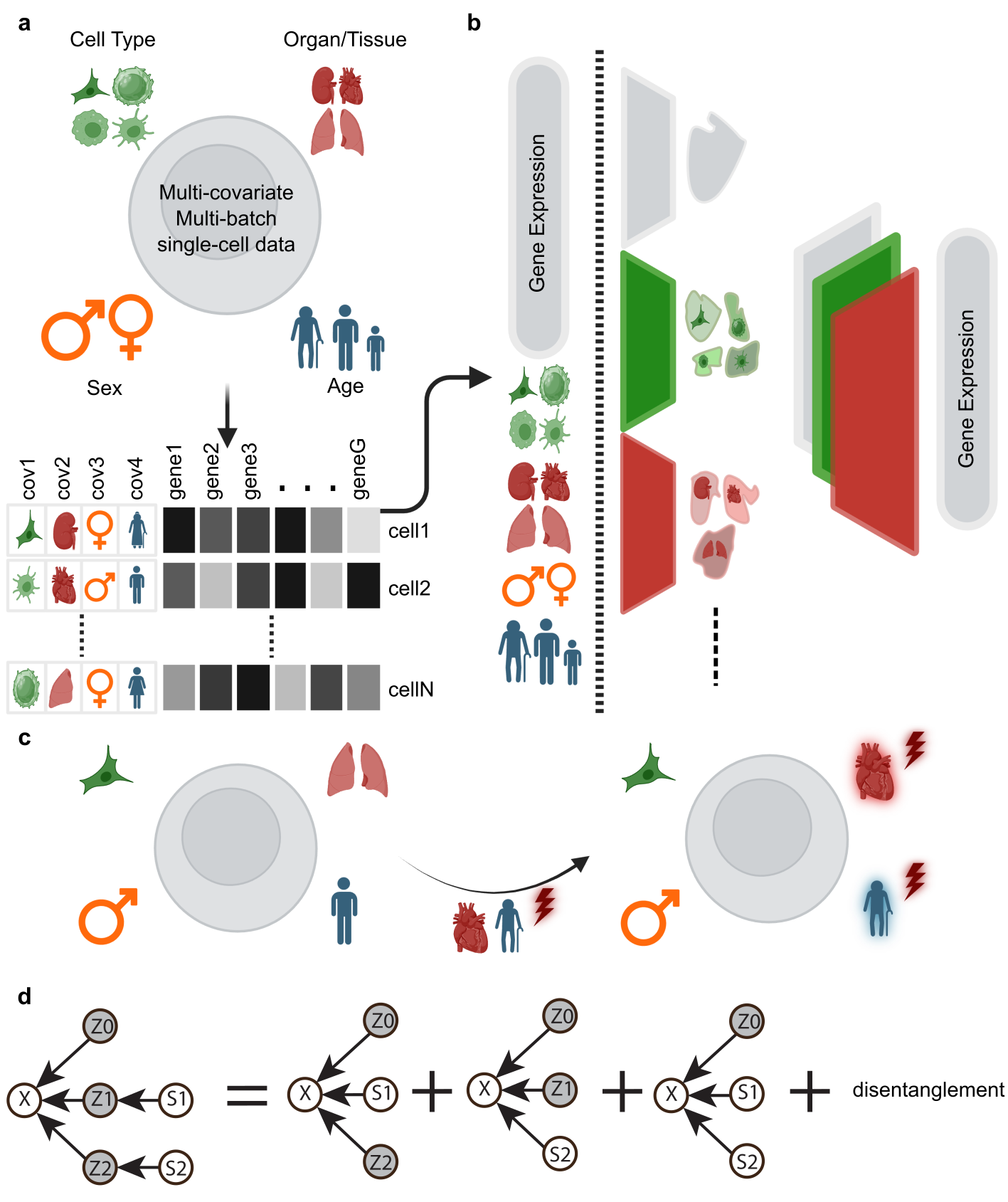
CellDISECT (Cell DISentangled Experts for Covariate counTerfactuals) is a powerful causal generative model that enhances single-cell analysis by:
- 🔍 Disentangling Variations: Separates covariate variations at test time
- 🧪 Counterfactual Predictions: Learns to make accurate counterfactual predictions
- 🎯 Flexible Fairness: Achieves flexible fairness through expert models for each latent space
- 🔬 Enhanced Discovery: Captures both covariate-specific information and novel biological insights
Visit our comprehensive documentation for:
- Detailed API reference
- Step-by-step tutorials
- Best practices and examples
- Advanced usage guides
We recommend using Anaconda/Miniconda. Create and activate a new environment:
conda create -n CellDISECT python=3.9
conda activate CellDISECT- Install PyTorch (tested with pytorch 2.1.2 and cuda 12):
conda install pytorch==2.1.2 torchvision==0.16.2 torchaudio==2.1.2 pytorch-cuda=12.1 -c pytorch -c nvidia- Install CellDISECT:
# Via pip (stable version)
pip install celldisect
# Or via GitHub (latest development version)
pip install git+https://github.com/Lotfollahi-lab/CellDISECTClick to expand optional installations
RAPIDS/rapids-singlecell:
pip install \
--extra-index-url=https://pypi.nvidia.com \
cudf-cu12==24.4.* dask-cudf-cu12==24.4.* cuml-cu12==24.4.* \
cugraph-cu12==24.4.* cuspatial-cu12==24.4.* cuproj-cu12==24.4.* \
cuxfilter-cu12==24.4.* cucim-cu12==24.4.* pylibraft-cu12==24.4.* \
raft-dask-cu12==24.4.* cuvs-cu12==24.4.*
pip install rapids-singlecellCUDA-enabled JAX:
pip install -U "jax[cuda12_pip]" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html| Tutorial | Description | Links |
|---|---|---|
| Basic Training | Learn how to train CellDISECT and make counterfactual predictions using the Kang dataset |  |
We welcome contributions! Please see our contributing guidelines for details on how to:
- Report issues
- Submit bug fixes
- Propose new features
- Submit pull requests
This project is licensed under the BSD 3-Clause License - see the LICENSE file for details.
For questions and support:
- Open an issue
- Visit our documentation
If you use CellDISECT in your research, please cite our paper:
Megas, S., Amani, A., Rose, A., Dufva, O., Shamsaie, K., Asadollahzadeh, H., Polanski, K., Haniffa, M., Teichmann, S. A., & Lotfollahi, M. (2025). Integrating multi-covariate disentanglement with counterfactual analysis on synthetic data enables cell type discovery and counterfactual predictions. bioRxiv. https://doi.org/10.1101/2025.06.03.657578
@article{Megas2025CellDISECT,
title={Integrating multi-covariate disentanglement with counterfactual analysis on synthetic data enables cell type discovery and counterfactual predictions},
author={Megas, Stathis and Amani, Arian and Rose, Antony and Dufva, Olli and Shamsaie, Kian and Asadollahzadeh, Hesam and Polanski, Krzysztof and Haniffa, Muzlifah and Teichmann, Sarah Amalia and Lotfollahi, Mohammad},
journal={bioRxiv},
year={2025},
doi={10.1101/2025.06.03.657578},
elocation-id={2025.06.03.657578},
publisher={Cold Spring Harbor Laboratory},
URL={https://www.biorxiv.org/content/10.1101/2025.06.03.657578v1}
}