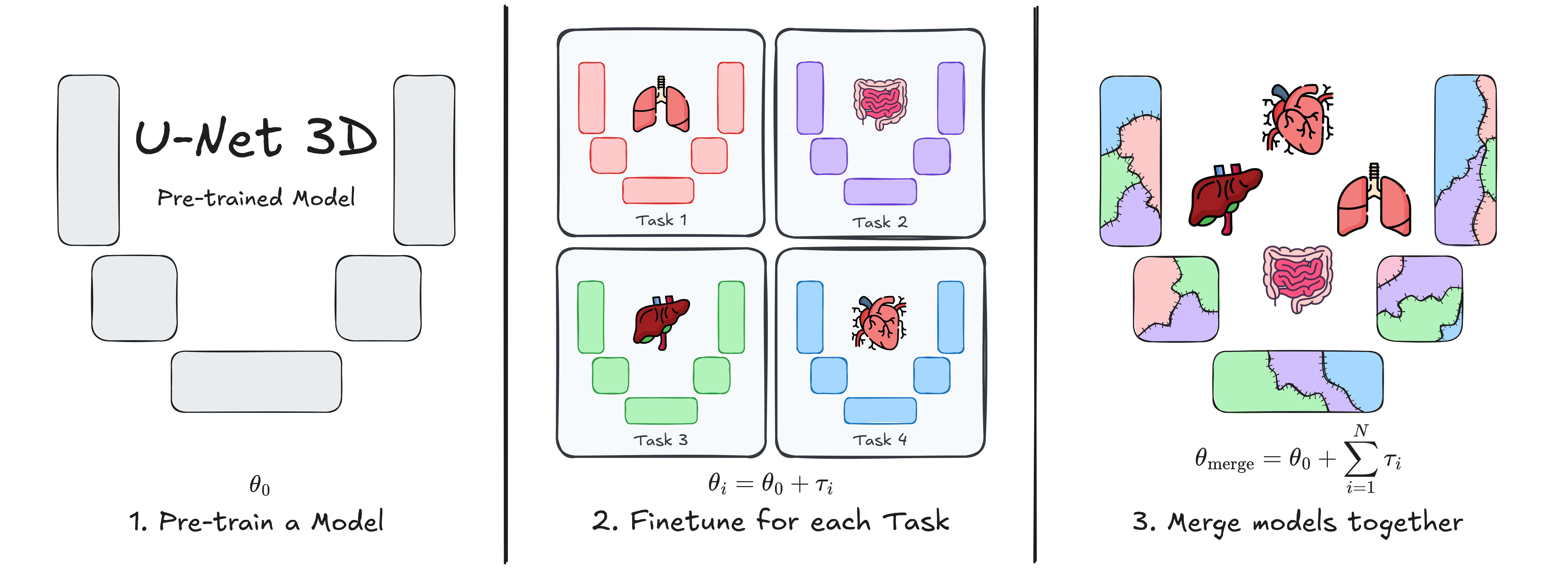
This repository contains the implementation of U-Net Transplant, a framework for efficient model merging in 3D medical image segmentation. Model merging enables the combination of specialized segmentation models without requiring full retraining, offering a flexible and privacy-conscious solution for updating AI models in clinical applications.
Our approach leverages task vectors and encourages wide minima during pre-training to enhance the effectiveness of model merging. We evaluate this method using the ToothFairy2 and BTCV Abdomen datasets with a standard 3D U-Net architecture, demonstrating its ability to integrate multiple specialized segmentation tasks into a single model.
Authors
Luca Lumetti*, Giacomo Capitani*, Elisa Ficarra, Costantino Grana, Simone Calderara, Angelo Porrello†, Federico Bolelli†
* Equal contributions
† Equal supervision
The related checkpoints and task vectors used in the paper will be available from the 23rd June 2025.
git clone [email protected]:LucaLumetti/UNetTransplant.git
cd UNetTransplantpython -m venv env
source env/bin/activate
pip install -r requirements.txtEnsure the datasets are downloaded and organized following the nnUNet dataset format.
- BTCV Abdomen: Download Here
- ToothFairy2: Download Here
- AMOS: Download Here
- ZhimingCui: Available upon request from the authors (Paper)
You can also download pretrained checkpoints and task vectors:
#!/bin/bash
BASE_ABDOMEN="https://huggingface.co/Lumett/UNetTransplant/resolve/main/Abdomen"
BASE_TOOTHFAIRY="https://huggingface.co/Lumett/UNetTransplant/resolve/main/ToothFairy"
abdomen_files=(
Pretrain_AMOS.pth
TaskVector_Kidney_Abdomen.pth
TaskVector_Liver_Abdomen.pth
TaskVector_Spleen_Abdomen.pth
TaskVector_Stomach_Abdomen.pth
)
toothfairy_files=(
Pretrain_Cui.pth
TaskVector_Canals_ToothFairy2.pth
TaskVector_Mandible_ToothFairy2.pth
TaskVector_Teeth_ToothFairy2.pth
TaskVector_Pharynx_ToothFairy2.pth
)
echo "🩻 Downloading Abdomen files..."
for file in "${abdomen_files[@]}"; do
wget -c "${BASE_ABDOMEN}/${file}"
done
echo "🦷 Downloading ToothFairy files..."
for file in "${toothfairy_files[@]}"; do
wget -c "${BASE_TOOTHFAIRY}/${file}"
doneThe main script for running experiments is main.py. It requires specifying the type of experiment and a configuration file that defines dataset, model, optimizer, and training parameters.
python main.py --experiment <EXPERIMENT_TYPE> --config <CONFIG_PATH> [--expname <NAME>] [--override <PARAMS>]-
--experiment: Specifies the type of experiment to run."PretrainExperiment"→ Pretrains the model from scratch."TaskVectorTrainExperiment"→ Trains a task vector using a pretrained checkpoint.
-
--config: Path to the configuration file, which defines dataset, model, and training settings. -
--expname(optional): Custom experiment name. If not provided, the config filename is used. -
--override(optional): Allows overriding config values at runtime. Example:python main.py --experiment PretrainExperiment --config configs/default.yaml --override DataConfig.BATCH_SIZE=4 OptimizerConfig.LR=0.01
The configuration file defines:
- Dataset (
DataConfig): Path, batch size, patch size, and datasets used. - Model (
BackboneConfig&HeadsConfig): Architecture, checkpoints, and initialization. - Optimizer (
OptimizerConfig): Learning rates, weight decay, and momentum. - Loss Function (
LossConfig): Defines the loss function used. - Training (
TrainConfig): Number of epochs, checkpoint saving, and resume options.
Check the provided configs for examples.
- Pretraining a model:
python main.py --experiment PretrainExperiment --config configs/miccai2025/pretrain_stable.yaml
- Training a task vector from a checkpoint:
python main.py --experiment TaskVectorTrainExperiment --config configs/miccai2025/finetune.yaml --override BackboneConfig.PRETRAIN_CHECKPOINTS="/path/to/checkpoint.pth"
For further details, refer to the config files used in our experiments under the configs folder.
If you used our work, please cite it:
@incollection{lumetti2025u,
title={U-Net Transplant: The Role of Pre-training for Model Merging in 3D Medical Segmentation},
author={Lumetti, Luca and Capitani, Giacomo and Ficarra, Elisa and Grana, Costantino and Calderara, Simone and Porrello, Angelo and Bolelli, Federico and others},
booktitle={Medical Image Computing and Computer Assisted Intervention--MICCAI 2025},
year={2025}
}