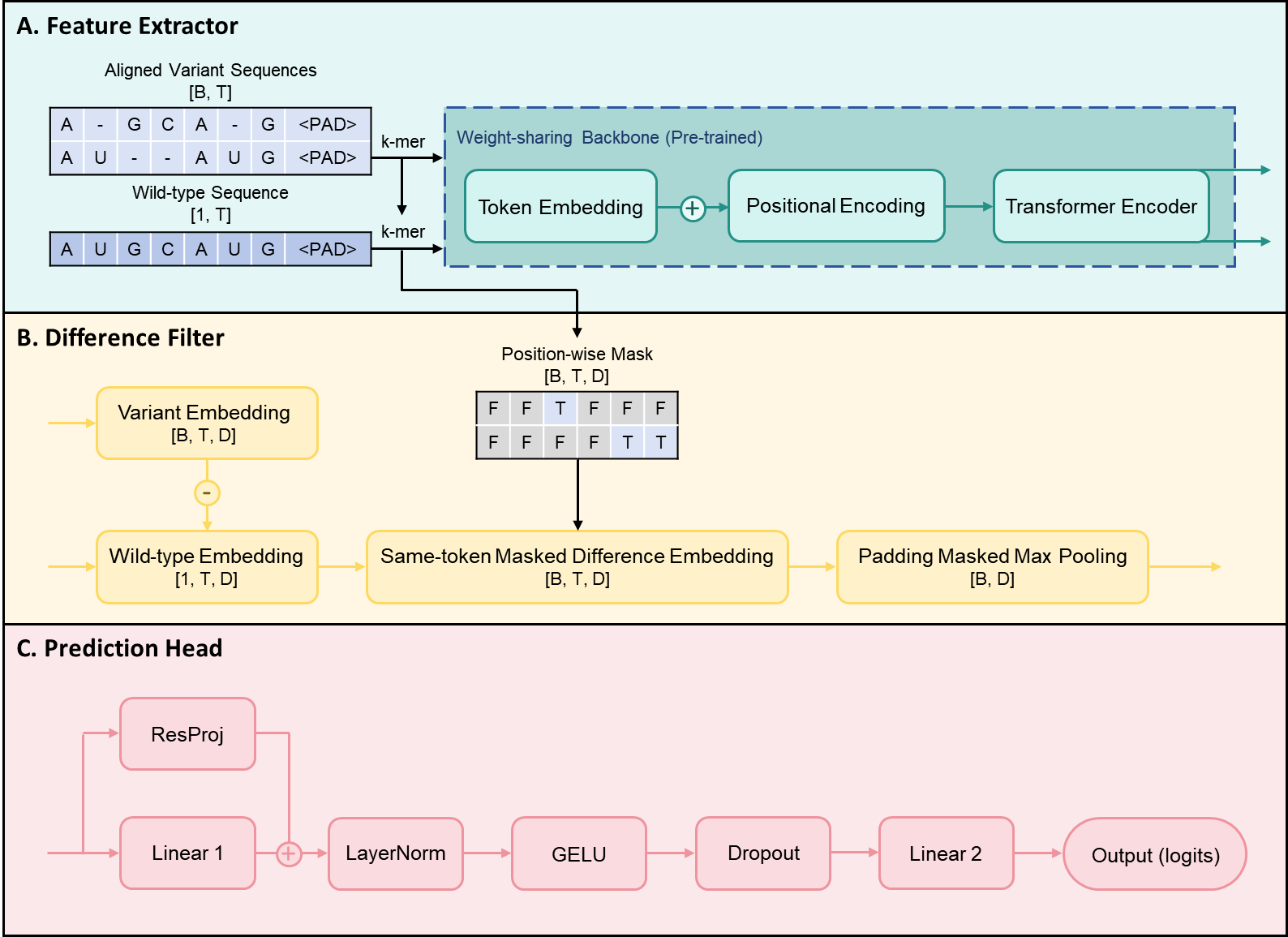
RDTransformer is a Transformer-based model designed to predict the functional effects of mutations in biological sequences (RNA, DNA, or proteins). Built upon a Transformer backbone, it introduces a dynamic difference-based embedding mechanism relative to the wild-type sequence. This mechanism filters out noise from non-mutational sites while emphasizing the effects of introduced or natural mutations.
The pretraining data was sourced from the RNAcentral database.
Citation:
RNAcentral Consortium.
RNAcentral 2021: secondary structure integration, improved sequence search and new member databases.
Nucleic Acids Research, 2021; 49(D1):D212-D220.
https://doi.org/10.1093/nar/gkaa921
Both raw and preprocessed pretraining datasets are available here.
The full implementation is available here.
1. Create environment:
conda env create -f environment.ymlNote: This is a CPU-only environment to maximize compatibility. The original experiments in the paper were conducted with the following configuration:
- PyTorch 2.6.0 + CUDA 12.4
- NVIDIA driver 555.42.06
- cuDNN: as bundled with the PyTorch 2.6.0+cu124 distribution
To reproduce GPU-accelerated results, install the matching CUDA-enabled PyTorch wheel:
pip install torch==2.6.0+cu124 --index-url https://download.pytorch.org/whl/cu124
2. Activate environment:
conda activate rdt_envConfigure finetuning using the YAML files in src/finetune_configs/, then run scripts from src/
1. Run pretraining:
python pretrain.py2. Run cross-validation for fine-tuning:
python finetune_cv.py --config finetune_configs/wb_cv_config.yaml
python finetune_cv.py --config finetune_configs/elisa_cv_config.yaml3. Run finetuning on full training set:
python finetune_fulltrain.py --config finetune_configs/wb_fulltrain_config.yaml
python finetune_fulltrain.py --config finetune_configs/elisa_fulltrain_config.yaml4. Run evaluation on held-out test set:
python finetune_test.py --config finetune_configs/wb_test_config.yaml
python finetune_test.py --config finetune_configs/elisa_test_config.yaml