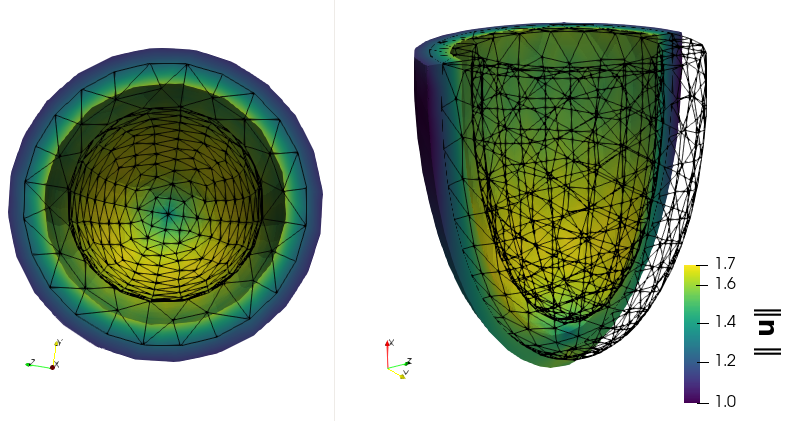
fenicsx-pulse is a cardiac mechanics solver based on FEniCSx. It is a successor of pulse which is a cardiac mechanics solver based on FEniCS.
- Documentation: https://finsberg.github.io/fenicsx-pulse/
- Source code: https://github.com/finsberg/fenicsx-pulse
You can install the library with pip
python3 -m pip install fenicsx-pulse
or with conda
conda install -c conda-forge fenicsx-pulse
Note that installing with pip requires FEniCSx already installed
We also provide a pre-built docker image with FEniCSx and fenicsx_pulse installed. You pull this image using the command
docker pull ghcr.io/finsberg/fenicsx-pulse:v0.5.1
Here is a minimal example of how to use fenicsx-pulse to solve a simple cardiac mechanics problem.
import numpy as np
import dolfinx
import cardiac_geometries
import pulse
# Create a geometry with cardiac-geometries
geo = cardiac_geometries.mesh.lv_ellipsoid(
outdir="geometry",
create_fibers=True,
fiber_space="Quadrature_6",
)
# Convert the geometry to a pulse.Geometry
geometry = pulse.HeartGeometry.from_cardiac_geometries(geo, metadata={"quadrature_degree": 6})
# Create a material model
material_params = pulse.HolzapfelOgden.transversely_isotropic_parameters()
material = pulse.HolzapfelOgden(f0=geo.f0, s0=geo.s0, **material_params)
# Define model for active contraction
Ta = pulse.Variable(dolfinx.fem.Constant(geometry.mesh, dolfinx.default_scalar_type(0.0)), "kPa")
active_model = pulse.ActiveStress(geo.f0, activation=Ta)
# Define mode for compressibility
comp_model = pulse.Incompressible()
# Assemble into a cardiac model
model = pulse.CardiacModel(
material=material,
active=active_model,
compressibility=comp_model,
)
# Define boundary conditions
traction = pulse.Variable(
dolfinx.fem.Constant(geometry.mesh, dolfinx.default_scalar_type(0.0)), "kPa"
)
neumann = pulse.NeumannBC(traction=traction, marker=geometry.markers["ENDO"][0])
def dirichlet_bc(V: dolfinx.fem.FunctionSpace):
# Find facets for the BASE marker
facets = geo.ffun.find(geo.markers["BASE"][0])
# Locate degrees of freedom for the x-component (sub(0)) on these facets
dofs = dolfinx.fem.locate_dofs_topological(V.sub(0), geo.mesh.topology.dim - 1, facets)
# Return the Dirichlet BC object
return [dolfinx.fem.dirichletbc(0.0, dofs, V.sub(0))]
robin_epi = pulse.RobinBC(
value=pulse.Variable(
dolfinx.fem.Constant(geometry.mesh, dolfinx.default_scalar_type(1e3)),
"Pa / m",
),
marker=geometry.markers["EPI"][0],
)
robin_base = pulse.RobinBC(
value=pulse.Variable(
dolfinx.fem.Constant(geometry.mesh, dolfinx.default_scalar_type(1e3)),
"Pa / m",
),
marker=geometry.markers["BASE"][0],
)
bcs = pulse.BoundaryConditions(neumann=(neumann,), dirichlet=(dirichlet_bc,), robin=(robin_base, robin_epi))
# Create a mechanics problem
problem = pulse.StaticProblem(
model=model,
geometry=geometry,
bcs=bcs,
)
# Perform an initial solve
problem.solve()
# Create a file for storing the solution
vtx = dolfinx.io.VTXWriter(geometry.mesh.comm, "displacement.bp", [problem.u], engine="BP4")
vtx.write(0.0)
# Assign a pressure and activation and ramp them up in steps
target_pressure = 5.0 # kPa
target_activation = 5.0 # kPa
num_steps = 5
for i, (pressure, activation) in enumerate(
zip(np.linspace(0, target_pressure, num_steps), np.linspace(0, target_activation, num_steps))
):
traction.assign(pressure) # kPa
Ta.assign(activation) # kPa
problem.solve()
# Save the displacement field
vtx.write(i + 1)
vtx.close()A more realistic example is visualized here:
u_simulation_warped.mp4
Checkout out the demos in the documentation for more examples.
See https://finsberg.github.io/fenicsx-pulse/CONTRIBUTING.html