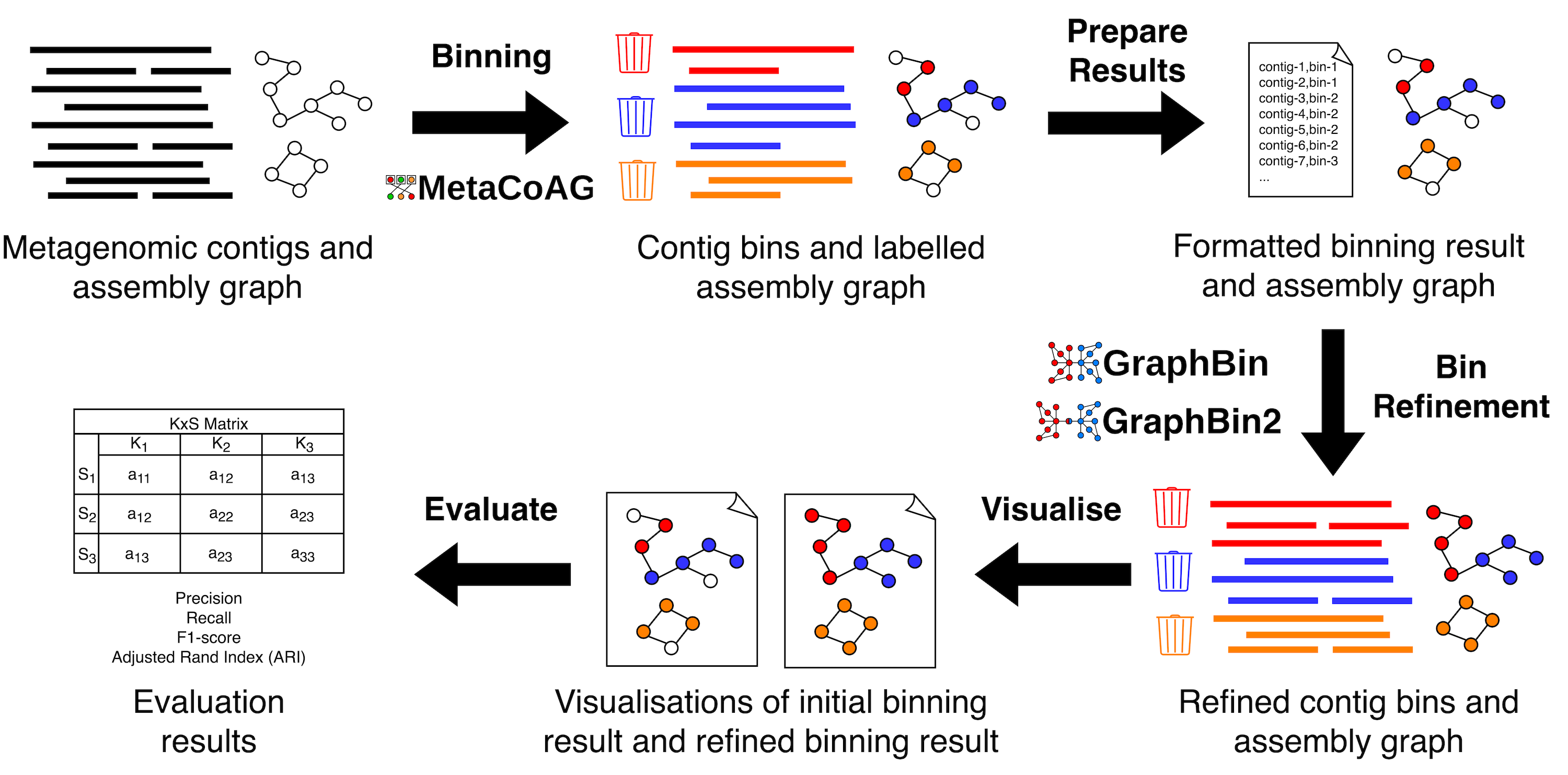
GraphBin-Tk combines assembly graph-based metagenomic bin-refinement and binning techniques GraphBin, GraphBin2 and MetaCoAG along with additional processing functionality to visualise and evaluate results, into one comprehensive toolkit. GraphBin-Tk enables users to seamlessly perform a wide range of tasks related to metagenomic binning and eliminates any compatibility issues that may arise from running separate tools. GraphBin-Tk is available on bioconda and PyPI.
For detailed instructions on installation and usage, please refer to the documentation hosted at Read the Docs.
You can install GraphBin-Tk using the bioconda distribution. You can download conda from
Anaconda or Miniconda. You can also use mamba instead of conda.
# add channels
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
# create conda environment
conda create -n gbintk
# activate conda environment
conda activate gbintk
# install gbintk
conda install -c bioconda gbintk
# check gbintk installation
gbintk --helpYou can install GraphBin-Tk using pip from the PyPI distribution.
# install gbintk
pip install gbintk
# check gbintk installation
gbintk --helpPlease follow the steps below to install gbintk using flit for development.
# clone repository
git clone https://github.com/metagentools/gbintk.git
# move to gbintk directory
cd gbintk
# create and activate conda env
conda env create -f environment.yml
conda activate gbintk
# install using flit
flit install -s --python `which python`
# test installation
gbintk --helpRun gbintk --help or gbintk -h to list the help message for GraphBin-Tk.
Usage: gbintk [OPTIONS] COMMAND [ARGS]...
gbintk (GraphBin-Tk): Assembly graph-based metagenomic binning toolkit
Options:
-v, --version Show the version and exit.
-h, --help Show this message and exit.
Commands:
graphbin GraphBin: Refined Binning of Metagenomic Contigs using...
graphbin2 GraphBin2: Refined and Overlapped Binning of Metagenomic...
metacoag MetaCoAG: Binning Metagenomic Contigs via Composition,...
prepare Format the initial binning result from an existing binning tool
visualise Visualise binning and refinement results
evaluate Evaluate the binning results given a ground truthPlease refer to DEMO.md for example code to run using test data provided. For further details on available subcommands and examples, please refer to the documentation hosted at Read the Docs.
GraphBin-Tk is currently under review at the Journal of Open Source Software.
If you use GraphBin-Tk in your work, please cite the relevant tools.
GraphBin
Vijini Mallawaarachchi, Anuradha Wickramarachchi, Yu Lin. GraphBin: Refined binning of metagenomic contigs using assembly graphs. Bioinformatics, Volume 36, Issue 11, June 2020, Pages 3307–3313, DOI: https://doi.org/10.1093/bioinformatics/btaa180
GraphBin2
Vijini G. Mallawaarachchi, Anuradha S. Wickramarachchi, and Yu Lin. GraphBin2: Refined and Overlapped Binning of Metagenomic Contigs Using Assembly Graphs. In 20th International Workshop on Algorithms in Bioinformatics (WABI 2020). Leibniz International Proceedings in Informatics (LIPIcs), Volume 172, pp. 8:1-8:21, Schloss Dagstuhl – Leibniz-Zentrum für Informatik (2020). DOI: https://doi.org/10.4230/LIPIcs.WABI.2020.8
Mallawaarachchi, V.G., Wickramarachchi, A.S. & Lin, Y. Improving metagenomic binning results with overlapped bins using assembly graphs. Algorithms Mol Biol 16, 3 (2021). DOI: https://doi.org/10.1186/s13015-021-00185-6
MetaCoAG
Mallawaarachchi, V., Lin, Y. (2022). MetaCoAG: Binning Metagenomic Contigs via Composition, Coverage and Assembly Graphs. In: Pe'er, I. (eds) Research in Computational Molecular Biology. RECOMB 2022. Lecture Notes in Computer Science(), vol 13278. Springer, Cham. DOI: https://doi.org/10.1007/978-3-031-04749-7_5
Vijini Mallawaarachchi and Yu Lin. Accurate Binning of Metagenomic Contigs Using Composition, Coverage, and Assembly Graphs. Journal of Computational Biology 2022 29:12, 1357-1376. DOI: https://doi.org/10.1089/cmb.2022.0262
Assemblers used to generate contigs and assembly graphs
Please cite the assembler used for metagenome assembly from the following list.
Sergey Nurk, Dmitry Meleshko, Anton Korobeynikov and Pavel A. Pevzner. metaSPAdes: a new versatile metagenomic assembler. Genome Research (2017) 27:5, 824-834. DOI: https://doi.org/10.1101/gr.213959.116
Dinghua Li, Chi-Man Liu, Ruibang Luo, Kunihiko Sadakane and Tak-Wah Lam. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:10 (2015), 1674-1676. DOI: https://doi.org/10.1093/bioinformatics/btv033
Mikhail Kolmogorov, Derek M. Bickhart, Bahar Behsaz, Alexey Gurevich, Mikhail Rayko, Sung Bong Shin, Kristen Kuhn, Jeffrey Yuan, Evgeny Polevikov, Timothy P. L. Smith and Pavel A. Pevzner. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nature Methods 17, (2020), 1103–1110. DOI: https://doi.org/10.1038/s41592-020-00971-x
Other software
Mina Rho, Haixu Tang and Yuzhen Ye. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Research, Volume 38, Issue 20, 1 November 2010, Page e191. DOI: https://doi.org/10.1093/nar/gkq747
Eddy, S.R. (2011) Accelerated Profile HMM Searches. PLOS Computational Biology 7(10): e1002195. DOI: https://doi.org/10.1371/journal.pcbi.1002195
Making Sense from Sequence — cogent3. URL: https://cogent3.org/
Csardi, G., & Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems, 1695. URL: https://python.igraph.org/
Aric A. Hagberg, Daniel A. Schult and Pieter J. Swart, “Exploring network structure, dynamics, and function using NetworkX”, in Proceedings of the 7th Python in Science Conference (SciPy2008), Gäel Varoquaux, Travis Vaught, and Jarrod Millman (Eds), (Pasadena, CA USA), pp. 11–15, Aug 2008. URL: https://networkx.org/
Harris, C.R., Millman, K.J., van der Walt, S.J. et al. Array programming with NumPy. Nature 585, 357–362 (2020). DOI: https://doi.org/10.1038/s41586-020-2649-2
Pauli Virtanen, Ralf Gommers, Travis E. Oliphant, Matt Haberland, Tyler Reddy, David Cournapeau, Evgeni Burovski, Pearu Peterson, Warren Weckesser, Jonathan Bright, Stéfan J. van der Walt, Matthew Brett, Joshua Wilson, K. Jarrod Millman, Nikolay Mayorov, Andrew R. J. Nelson, Eric Jones, Robert Kern, Eric Larson, CJ Carey, İlhan Polat, Yu Feng, Eric W. Moore, Jake VanderPlas, Denis Laxalde, Josef Perktold, Robert Cimrman, Ian Henriksen, E.A. Quintero, Charles R Harris, Anne M. Archibald, Antônio H. Ribeiro, Fabian Pedregosa, Paul van Mulbregt, and SciPy 1.0 Contributors. (2020) SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods, 17(3), 261-272. DOI: https://doi.org/10.1038/s41592-019-0686-2
GraphBin-Tk is funded by an Essential Open Source Software for Science Grant from the Chan Zuckerberg Initiative.