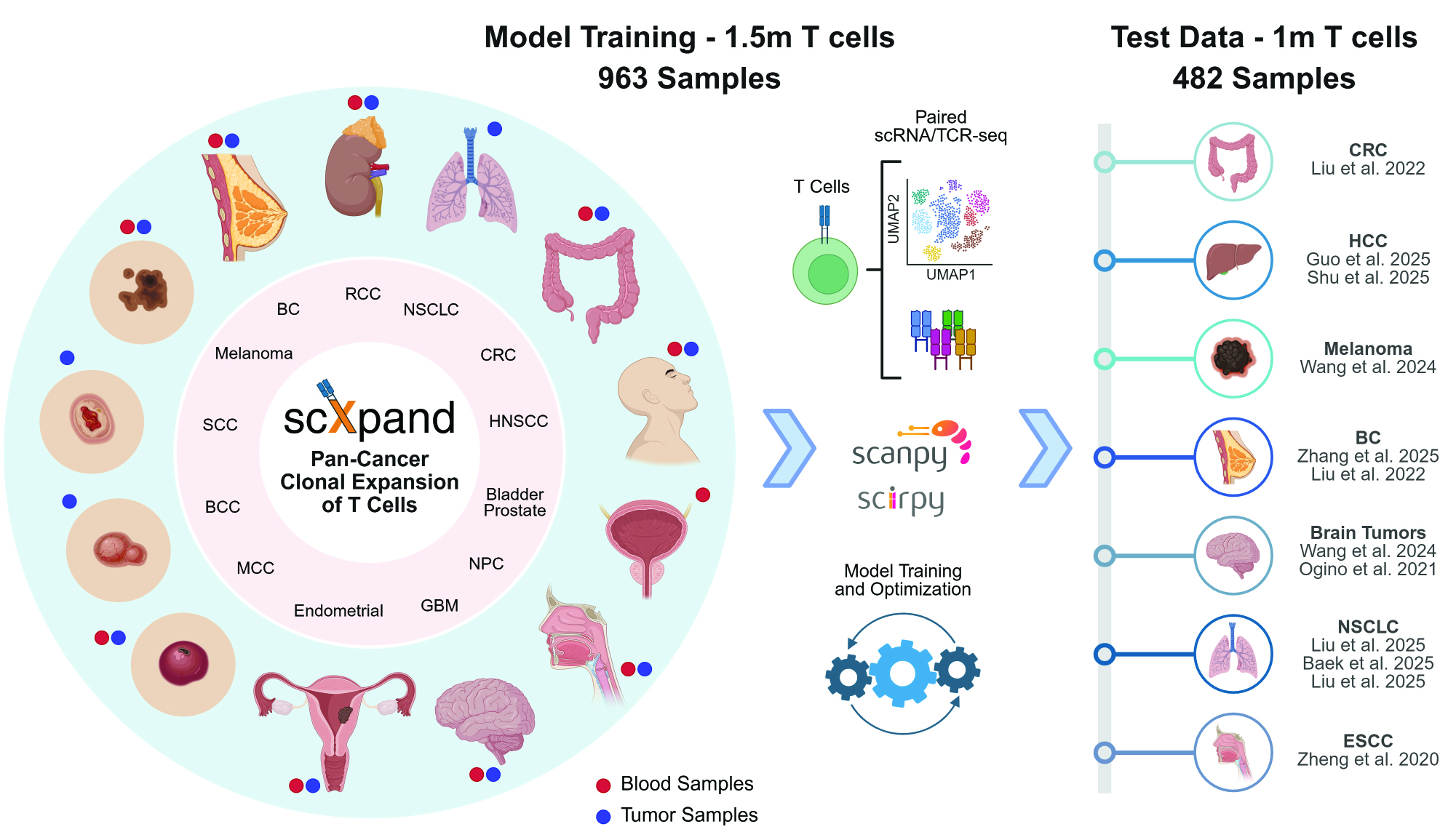
Detect T-cell clonal expansion from single-cell RNA sequencing data without paired TCR sequencing
Preprint • Documentation • Installation • Quick Start • Usage Guide • Citation
A framework for predicting T-cell clonal expansion from single-cell RNA sequencing data.
Manuscript in preparation - detailed methodology and benchmarks coming soon.
View full documentation for comprehensive guides and API reference.
- Multiple Model Architectures:
- Autoencoder-based: Encoder-decoder with reconstruction and classification heads
- MLP: Multi-layer perceptron
- LightGBM: Gradient boosted decision trees
- Linear Models: Logistic regression and support vector machines
- Scalable Processing: Handles millions of cells with memory-efficient data streaming from disk during training
- Automated Hyperparameter Optimization: Built-in Optuna integration for model tuning
For detailed installation instructions, please refer to our Installation Guide.
CUDA version (NVIDIA GPU):
With pip:
pip install --upgrade scxpand-cuda --extra-index-url https://download.pytorch.org/whl/cu128With uv:
uv pip install --upgrade scxpand-cuda --extra-index-url https://download.pytorch.org/whl/cu128 --index-strategy unsafe-best-matchCPU/Apple Silicon/Other GPUs:
With pip:
pip install --upgrade scxpandWith uv:
uv pip install --upgrade scxpandSee the Installation Guide
import scxpand
# Make sure that "your_data.h5ad" includes only T cells for the results to be meaningful
# Ensure that "your_data.var_names" are provided as Ensembl IDs (as the pre-trained models were trained using this gene representation)
# Please refer to our documentation for more information
# List available pre-trained models
scxpand.list_pretrained_models()
# Run inference with automatic model download
results = scxpand.run_inference(
model_name="pan_cancer_autoencoder", # default model
data_path="your_data.h5ad"
)
# Access predictions
predictions = results.predictions
if results.has_metrics:
print(f"AUROC: {results.get_auroc():.3f}")See our Tutorial Notebook for a complete example with data preprocessing, T-cell filtering, gene ID conversion, and model application using a real breast cancer dataset.
Setup & Getting Started:
- Installation Guide - Setup for local development of scXpand
- User Guide - Quick start and comprehensive workflow guide
- Data Format - Input data requirements and specifications
Using Pre-trained Models:
- Model Inference - Run predictions on new data with pre-trained models
Training Your Own Models:
- Model Training - Train models with CLI and programmatic API
- Hyperparameter Optimization - Automated model tuning with Optuna
Understanding Results:
- Model Architectures - Detailed architecture descriptions and configurations
- Evaluation Metrics - Performance assessment and interpretation
- Output Format - Understanding model outputs and results
📖 Full Documentation - Complete guides, API reference, and interactive tutorials
This project is licensed under the MIT License – see the LICENSE file for details.
If you use scXpand in your research, please cite:
Shorer, O., Amit, R., and Yizhak, K. (2025). scXpand: Pan-cancer detection of T-cell clonal expansion from single-cell RNA sequencing without paired single-cell TCR sequencing. Preprint at bioRxiv, https://doi.org/10.1101/2025.09.14.676069.
BibTeX
@article{shorer2025scxpand,
title={scXpand: Pan-cancer detection of T-cell clonal expansion from single-cell RNA sequencing without paired single-cell TCR sequencing},
author={Shorer, Ofir and Amit, Ron and Yizhak, Keren},
year={2025},
journal={bioRxiv},
doi={https://doi.org/10.1101/2025.09.14.676069}
}