Vgrid
DGGS and Cell-based Geocoding Utilities
Vgrid User Guide
- Using pip:
pip install vgrid --upgrade
- Vgrid Home: Vgrid DGGS
Convert lat, long in WGS84 CRS to DGGS cellID (H3, S2, rHEALPix, OpenEaggr ISEA4T and ISEA3H (Windows only), EASE-DGGS, QTM, OLC/ OpenLocationCode/ Google Plus Code, Geohash, GEOREF, MGRS, Tilecode, Quadkey, Maidenhead, GARS).
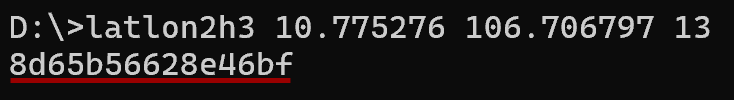
> latlon2h3 10.775276 106.706797 13 # latlon2h3 <lat> <lon> <resolution> [0..15]
> latlon2s2 10.775276 106.706797 21 # latlon2s2 <lat> <lon> <resolution> [0..30]
> latlon2rhealpix 10.775276 106.706797 14 # latlon2rhealpix <lat> <lon> <resolution> [1..15]
> latlon2isea4t 10.775276 106.706797 21 # latlon2isea4t <lat> <lon> <resolution> [0..39]
> latlon2isea3h 10.775276 106.706797 27 # latlon2isea3h <lat> <lon> <resolution> [0..40]
> latlon2ease 10.775276 106.706797 6 # latlon2easedggs <lat> <lon> <resolution> [0..6]
> latlon2dggrid 10.775276 106.706797 FULLER4D 10 # Linux only: latlon2dggrid <lat> <lon> <dggs_type> <resolution>
> latlon2qtm 10.775276 106.706797 11 # latlon2qtm <lat> <lon> <resolution> [1..24]
> latlon2olc 10.775276 106.706797 11 # latlon2olc <lat> <lon> <resolution> [2,4,6,8,10,11..15]
> latlon2geohash 10.775276 106.706797 9 # latlon2geohash <lat> <lon> <resolution>[1..10]
> latlon2georef 10.775276 106.706797 4 # latlon2georef <lat> <lon> <resolution> [0..5]
> latlon2mgrs 10.775276 106.706797 4 # latlon2mgrs <lat> <lon> <resolution> [0..5]
> latlon2tilecode 10.775276 106.706797 23 # latlon2tilecode <lat> <lon> <resolution> [0..29]
> latlon2quadkey 10.775276 106.706797 23 # latlon2quadkey <lat> <lon> <resolution> [0..29]
> latlon2maidenhead 10.775276 106.706797 4 # latlon2maidenhead <lat> <lon> <resolution> [1..4]
> latlon2gars 10.775276 106.706797 1 # latlon2gars <lat> <lon> <resolution> [1..4] (30,15,5,1 minutes)Convert DGGS cell ID to GeoJSON.
{"type": "FeatureCollection", "features": [{"type": "Feature", "geometry": {"type": "Polygon", "coordinates": [[[106.70789209957029, 10.77601109480422], [106.70745212203926, 10.777918172217145], [106.7055792173179, 10.778494886227104], [106.70414629547702, 10.777164500398262], [106.70458630070586, 10.775257408234628], [106.70645920007833, 10.774680716650128], [106.70789209957029, 10.77601109480422]]]}, "properties": {"h3": "8965b56628fffff", "resolution": 9, "center_lat": 10.7765878, "center_lon": 106.7060192, "avg_edge_len": 215.295, "cell_area": 120421.396}}]}
> h32geojson 8d65b56628e46bf
> s22geojson 31752f45cc94
> rhealpix2geojson R31260335553825
> isea4t2geojson 13102313331320133331133 # Windows only
> isea3h2geojson 1327916769,-55086 # Windows only
> ease2geojson L6.165767.02.02.22.45.63.05
> dggrid2geojson 8478420 FULLER4D 10 # Linux only: dggrid2geojson <DGGRID code in SEQNUM format> <DGGS Type> <resolution>
# <DGGS Type> chosen from [SUPERFUND,PLANETRISK,ISEA3H,ISEA4H,ISEA4T,ISEA4D,ISEA43H,ISEA7H,IGEO7,FULLER3H,FULLER4H,FULLER4T,FULLER4D,FULLER43H,FULLER7H]
> qtm2geojson 42012323131
> olc2geojson 7P28QPG4+4P7
> geohash2geojson w3gvk1td8
> georef2geojson VGBL42404651
> mgrs2geojson 34TGK56063228
> gzd # Create Grid Zone Designators - used by MGRS
> tilecode2geojson z23x6680749y3941729
> quadkey2geojson 13223011131020212310000
> maidenhead2geojson OK30is46
> gars2geojson 574JK1918Convert Vector layers (Point/ Multipoint, Linestring/ Multilinestring, polygon/ Multipolygon) in GeoJSON to DGGS.
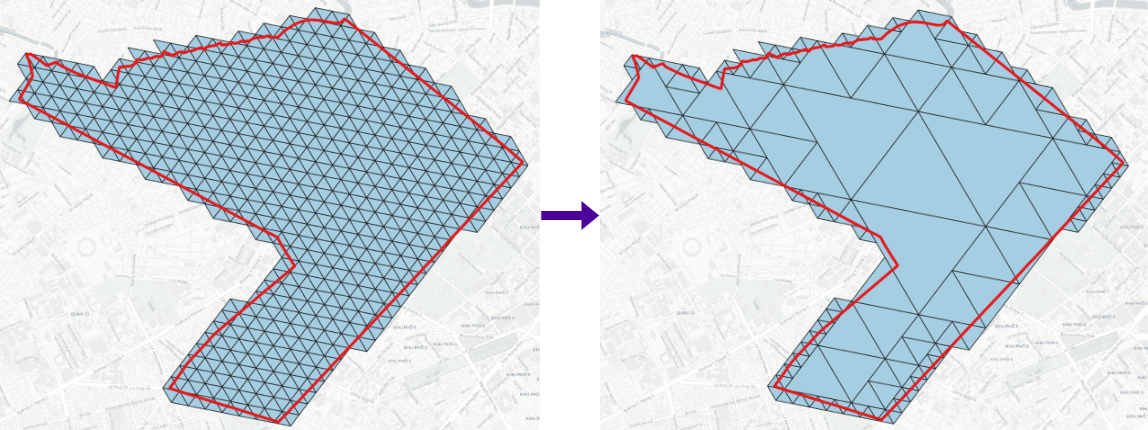
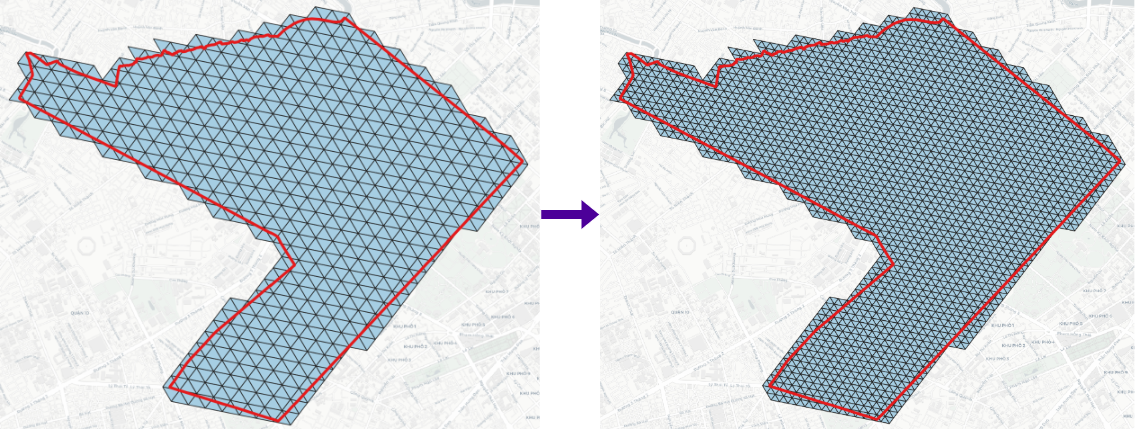
- Uncompact:
- Compact:
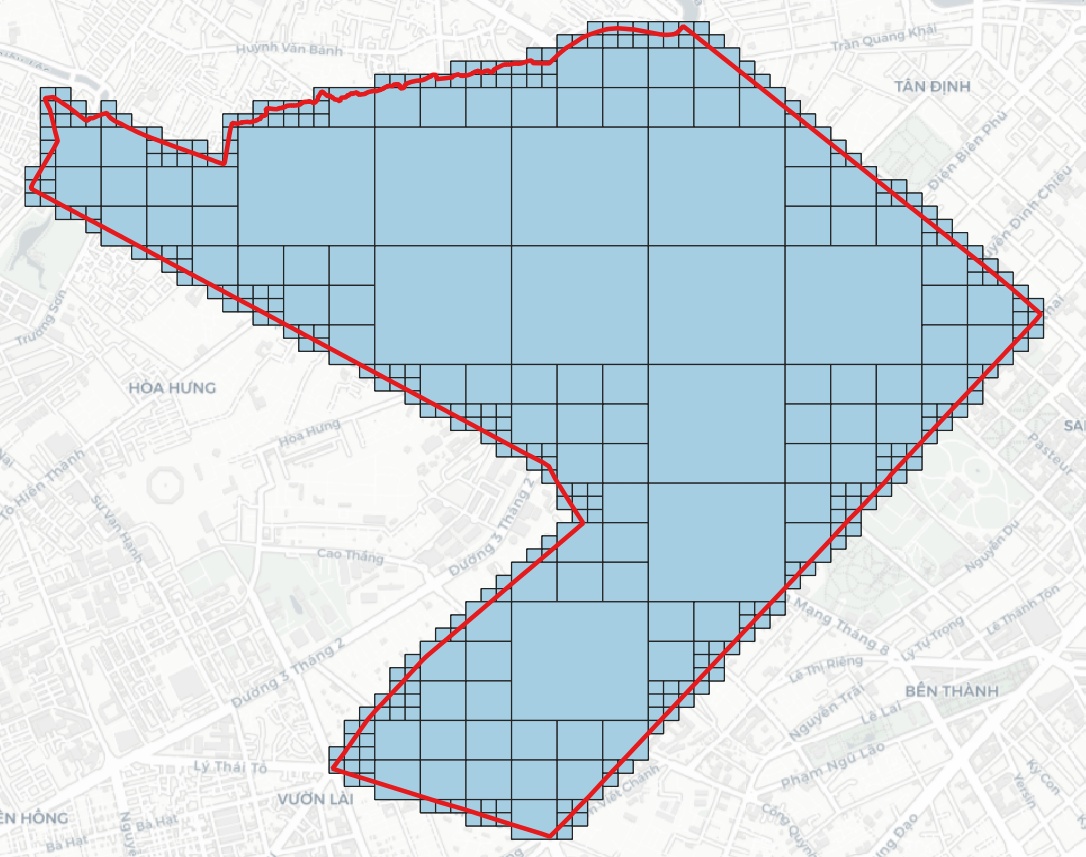
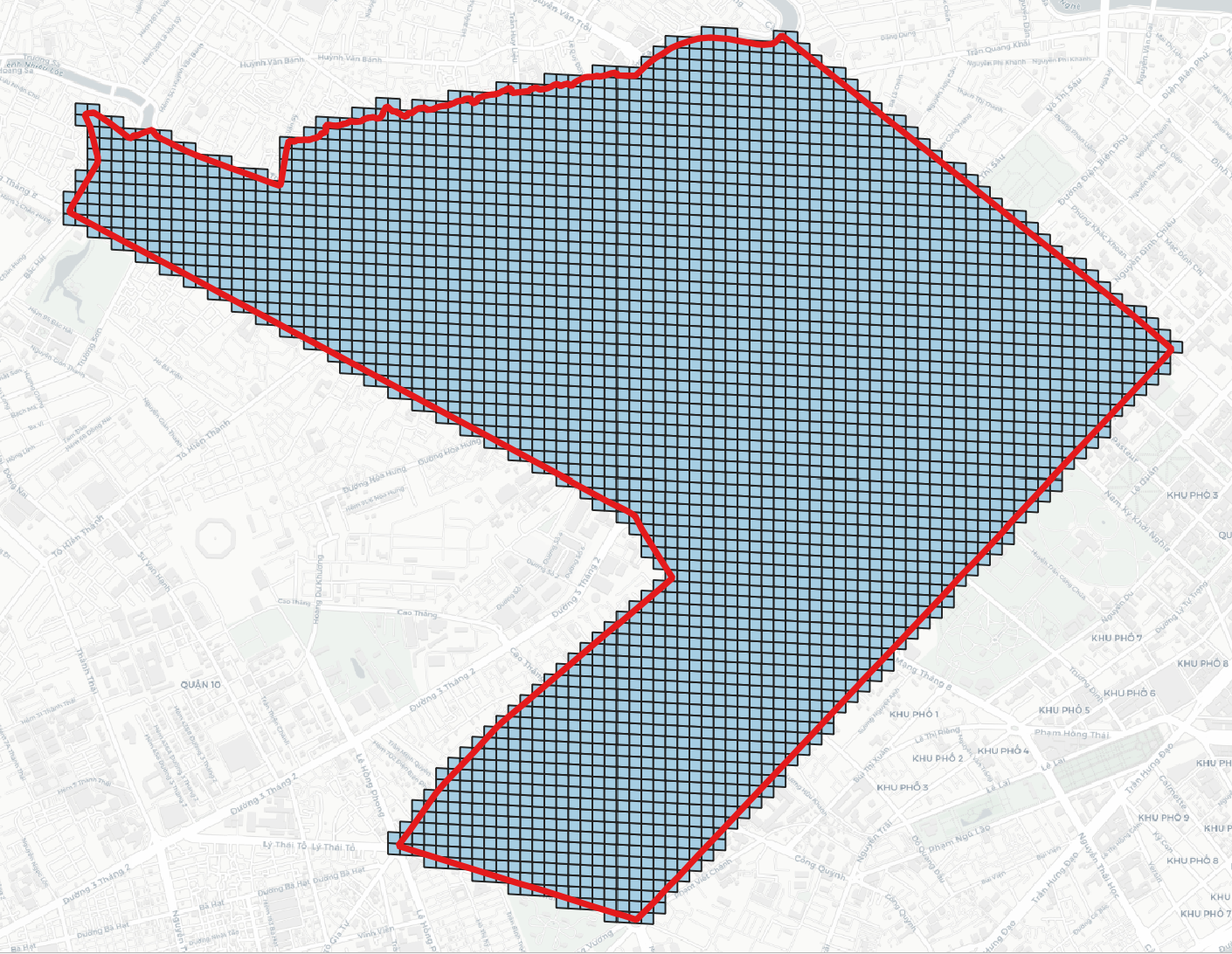
> geojson2h3 -r 11 -geojson polygon.geojson # geojson2h3 -r <resolution>[0..15] -geojson <GeoJSON file> -compact [optional]
> geojson2s2 -r 18 -geojson polygon.geojson -compact # geojson2s2 -r <resolution>[0..30] -geojson <GeoJSON file> -compact [optional]
> geojson2rhealpix -r 11 -geojson polygon.geojson # geojson2rhealpix -r <resolution>[1..15] -geojson <GeoJSON file> -compact [optional]
> geojson2isea4t -r 17 -geojson polygon.geojson # geojson2isea4t -r <resolution>[0..25] -geojson <GeoJSON file> -compact [optional]
> geojson2isea3h -r 18 -geojson polygon.geojson # geojson2isea3h -r <resolution>[1..32] -geojson <GeoJSON file> -compact [optional]
> geojson2ease -r 4 -geojson polygon.geojson # geojson2ease -r <resolution>[0..6] -geojson <GeoJSON file> -compact [optional]
> geojson2dggrid -t ISEA4H -r 17 -geojson polyline.geojson # Linux only: geojson2dggrid -t <DGGS Type> -r <resolution> -a <address_type, default is SEQNUM> -geojson <GeoJSON path>
# <Address Type> chosen from [Q2DI,SEQNUM,INTERLEAVE,PLANE,Q2DD,PROJTRI,VERTEX2DD,AIGEN,Z3,Z3_STRING,Z7,Z7_STRING,ZORDER,ZORDER_STRING]
> geojson2qtm -r 18 -geojson polygon.geojson # geojson2qtm -r <resolution>[1..24] -geojson <GeoJSON file> -compact [optional]
> geojson2olc -r 10 -geojson polygon.geojson # geojson2olc -r <resolution>[2,4,6,8,10,11..15] -geojson <GeoJSON file> -compact [optional]
> geojson2geohash -r 7 -geojson polygon.geojson # geojson2geohash -r <resolution>[1..10] -geojson <GeoJSON file> -compact [optional]
> geojson2mgrs -r 3 -geojson polygon.geojson # geojson2mgrs -r <resolution>[0..5] -geojson <GeoJSON file>
> geojson2tilecode -r 18 -geojson polygon.geojson # geojson2tilecode -r <resolution>[0..29] -geojson <GeoJSON file> -compact [optional]
> geojson2quadkey -r 18 -geojson polygon.geojson # geojson2quadkey -r <resolution>[0..29] -geojson <GeoJSON file> -compact [optional]Convert raster layers in geographic CRS to output DGGS with closest matching resolution.
> raster2h3 -raster raster.tif # raster2h3 -raster <raster in geographic CRS> -r <resolution>[0..15] [optional, defaults to the H3 resolution nearest to the raster's cell size]
> raster2s2 -raster raster.tif # raster2s2 -raster <raster in geographic CRS> -r <resolution>[0..24] [optional, defaults to the S2 resolution nearest to the raster's cell size]
> raster2rhealpix -raster raster.tif # raster2rhealpix -raster <raster in geographic CRS> -r <resolution>[0..15] [optional, defaults to the rHEALPix resolution nearest to the raster's cell size]
> raster2isea4t -raster raster.tif # Windows only: raster2isea4t -raster <raster in geographic CRS> -r <resolution>[0..23] [optional, defaults to the ISEA4T resolution nearest to the raster's cell size]
> raster2qtm -raster raster.tif # raster2qtm -raster <raster in geographic CRS> -r <resolution>[1..24] [optional, defaults to the QTM resolution nearest to the raster's cell size]
> raster2olc -raster raster.tif # raster2olc -raster <raster in geographic CRS> -r <resolution>[10..12] [optional, defaults to the OLC resolution nearest to the raster's cell size]
> raster2geohash -raster raster.tif # raster2geohash -raster <raster in geographic CRS> -r <resolution>[1..10] [optional, defaults to the Geohash resolution nearest to the raster's cell size]
> raster2tilecode -raster raster.tif # raster2tilecode -raster <raster in geographic CRS> -r <resolution>[0..26] [optional, defaults to the Tilecode resolution nearest to the raster's cell size]
> raster2quadkey -raster raster.tif # raster2quadkey -raster <raster in geographic CRS> -r <resolution>[0..26] [optional, defaults to the Quadkey resolution nearest to the raster's cell size]> h3compact -geojson h3.geojson -cellid h3 # h3compact -geojson <H3 in GeoJSON> -cellid [optional, 'h3' by default]
> s2compact -geojson s2.geojson -cellid s2 # s2compact -geojson <S2 in GeoJSON> -cellid [optional, 's2' by default]
> rhealpixcompact -geojson rhealpix.geojson -cellid rhealpix # rhealpixcompact -geojson <rHEALPix in GeoJSON> -cellid [optional, 'rhealpix' by default]
> isea4tcompact -geojson isea4t.geojson -cellid isea4t # Windows only: isea4tcompact -geojson <ISEA4T in GeoJSON> -cellid [optional, 'isea4t' by default]
> isea3hcompact -geojson isea3h.geojson -cellid isea3h # Windows only: isea3hcompact -geojson <ISEA3H in GeoJSON> -cellid [optional, 'isea3h' by default]
> easecompact -geojson ease.geojson -cellid ease # easecompact -geojson <EASE in GeoJSON> -cellid [optional, 'ease' by default]
> qtmcompact -geojson qtm.geojson -cellid qtm # qtmcompact -geojson <QTM in GeoJSON> -cellid [optional, 'qtm' by default]
> olccompact -geojson olc.geojson -cellid olc # olccompact -geojson <OLC in GeoJSON> -cellid [optional, 'olc' by default]
> geohashcompact -geojson geohash.geojson -cellid geohash # geohashcompact -geojson <Geohash in GeoJSON> -cellid [optional, 'geohash' by default]
> tilecodecompact -geojson tilecode.geojson -cellid tilecode # tilecodecompact -geojson <Tilecode in GeoJSON> -cellid [optional, 'tilecode' by default]
> quadkeycompact -geojson quadkey.geojson -cellid quadkey # quadkeycompact -geojson <quadkey in GeoJSON> -cellid [optional, 'quadkey' by default]> h3expand -geojson h3_11.geojson -r 12 -cellid h3 # h3expand -geojson <H3 in GeoJSON> -r <higher resolution>[0..15] -cellid [optional, 'h3' by default]
> s2expand -geojson s2_18.geojson -r 19 -cellid s2 # s2expand -geojson <S2 in GeoJSON> -r <higher resolution>[0..30] -cellid [optional, 's2' by default]
> rhealpixexpand -geojson rhealpix_10.geojson -r 11 -cellid rhealpix # rhealpixexpand -geojson <rHEALPix in GeoJSON> -r <higher resolution>[0..15] -cellid [optional, 'rhealpix' by default]
> isea4texpand -geojson isea4t_18.geojson -r 19 -cellid isea4t # Windows only: isea4texpand -geojson <ISEA4T in GeoJSON> -r <higher resolution>[0..25] -cellid [optional, 'isea4t' by default]
> isea3hexpand -geojson isea3h_18.geojson -r 19 -cellid isea3h # Windows only: isea3hexpand -geojson <ISEA3H in GeoJSON> -r <higher resolution>[0..25] -cellid [optional, 'isea3h' by default]
> easeexpand -geojson ease_4.geojson -r 5 -cellid ease # easeexpand -geojson <EASE in GeoJSON> -r <higher resolution>[0..6] -cellid [optional, 'ease' by default]
> qtmexpand -geojson qtm_18.geojson -r 19 -cellid qtm # qtmexpand -geojson <QTM in GeoJSON> -r <higher resolution>[1..24] -cellid [optional, 'qtm' by default]
> olcexpand -geojson olc_8.geojson -r 10 -cellid olc # olcexpand -geojson <OLC in GeoJSON> -r <higher resolution>[2,4,6,8,10..15] -cellid [optional, 'olc' by default]
> geohashexpand -geojson geohash_7.geojson -r 8 -cellid geohash # geohashexpand -geojson <Geohash in GeoJSON> -r <higher resolution>[1..10] -cellid [optional, 'geohash' by default]
> tilecodeexpand -geojson tilecode_18.geojson -r 19 -cellid tilecode # tilecodeexpand -geojson <Tilecode in GeoJSON> -r <higher resolution>[0..29] -cellid [optional, 'tilecode' by default]
> quadkeyexpand -geojson quadkey_18.geojson -r 19 -cellid quadkey # quadkeyexpand -geojson <Quadkey in GeoJSON> -r <higher resolution>[0..29] -cellid [optional, 'quadkey' by default]> csv2h3 h3.csv # Convert CSV with 'h3' column to H3 cells.
> csv2s2 s2.csv # Convert CSV with 's2' column to S2 cells.
> csv2rhealpix rhealpix.csv # Convert CSV with 'rhealpix' column to rHEALPix cells.
> csv2isea4t isea4t.csv # Windows only: Convert CSV with 'rhealpisea4t' column to ISEA4T cells.
> csv2isea3h isea3h.csv # Windows only: Convert CSV with 'isea3h' column to ISEA3H cells.
> csv2sease ease.csv # Convert CSV with 'ease' column to EASE cells.
> csv2qtm qtm.csv # Convert CSV with 'qtm' column to QTM cells.
> csv2olc olc.csv # Convert CSV with 'olc' column to OLC cells.
> csv2geohash geohash.csv # Convert CSV with 'geohash' column to Geohash cells.
> csv2georef georef.csv # Convert CSV with 'georef' column to GEOREF cells.
> csv2mgrs mgrs.csv # Convert CSV with 'mgrs' column to MGRS cells.
> csv2tilecode tilecode.csv # Convert CSV with 'tilecode' column to Tilecode cells.
> csv2quadkey quadkey.csv # Convert CSV with 'quadkey' column to Tilecode cells.
> csv2maidenhead maidenhead.csv # Convert CSV with 'maidenhead' column to Maidenhead cells.
> csv2gars gars.csv # Convert CSV with 'gars' column to GARS cells.Binning point layer to DGGS
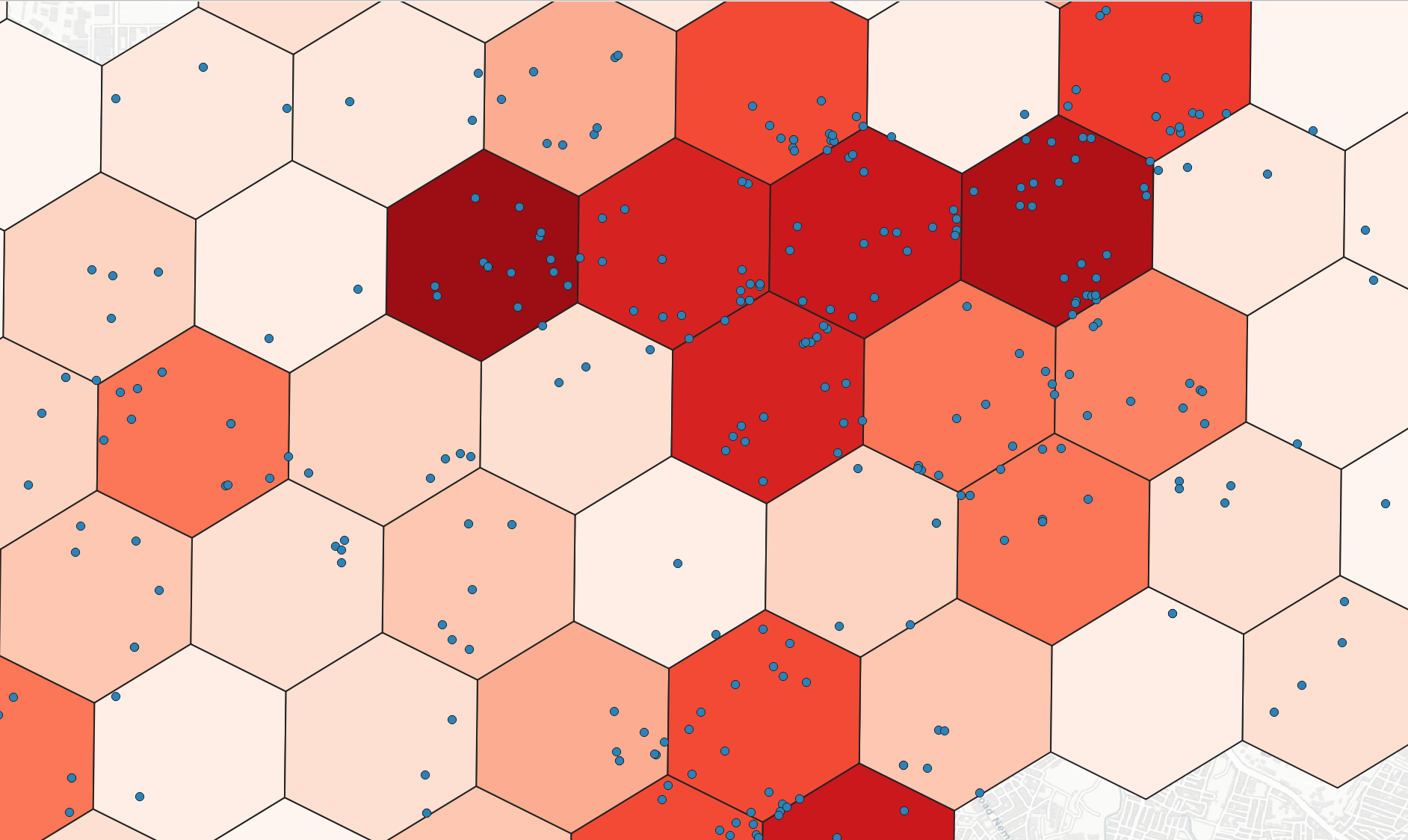
> h3bin -point point.geojson -r 8 -stats count -field numeric_field -category group # h3bin -point <point GeoJSON file> -r <resolution[0..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> s2bin -point point.geojson -r 13 -stats count -field numeric_field -category group # s2bin -point <point GeoJSON file> -r <resolution[0..30]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> rhealpixbin -point point.geojson -r 8 -stats count -field numeric_field -category group # rhealpixbin -point <point GeoJSON file> -r <resolution[0..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> isea4tbin -point point.geojson -r 13 -stats count -field numeric_field -category group # Windows only: isea4tbin -point <point GeoJSON file> -r <resolution[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> qtmbin -point point.geojson -r 13 -stats count -field numeric_field -category group # qtmbin -point <point GeoJSON file> -r <resolution[1..24]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> olcbin -point point.geojson -r 9 -stats count -field numeric_field -category group # olcbin -point <point GeoJSON file> -r <resolution[2,4,6,8,10..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> geohashbin -point point.geojson -r 6 -stats count -field numeric_field -category group # geohashbin -point <point GeoJSON file> -r <resolution[1..10]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> tilecodebin -point point.geojson -r 15 -stats count -field numeric_field -category group # tilecodebin -point <point GeoJSON file> -r <resolutin[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> quadkeybin -point point.geojson -r 13 -stats count -field numeric_field -category group # Windows only: quadkeybin -point <point GeoJSON file> -r <resolutin[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping] Resample input DGGS to output DGGS with closest matching resolution.
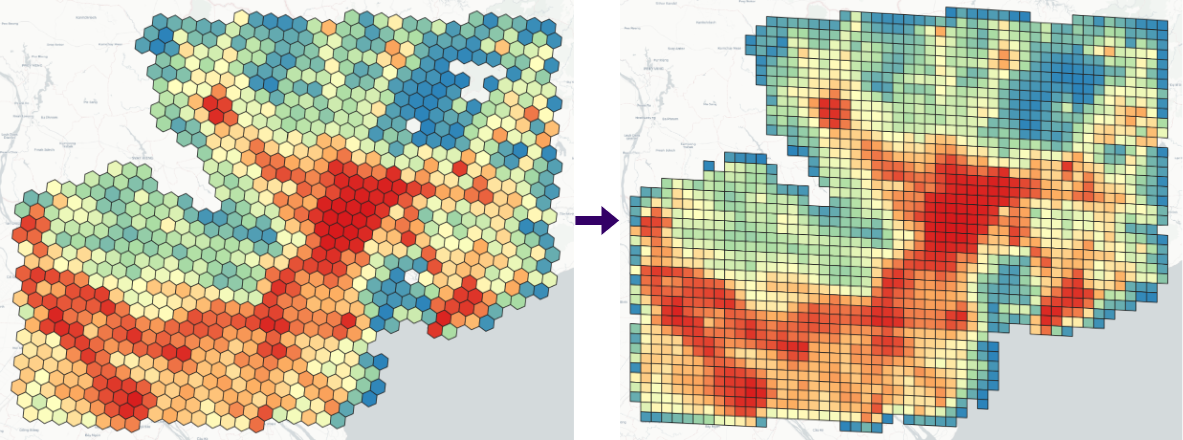
> resample -geojson vn_pop_h3_6.geojson -fromdggs h3 -todggs s2 -resamplefield population # resample -geojson <Input DGGS> -fromdggs <Input DGGS type> -todggs <Output DGGS Type> -resamplefield [Optional, Numeric field for resampling]Generate DGGS within a bounding box
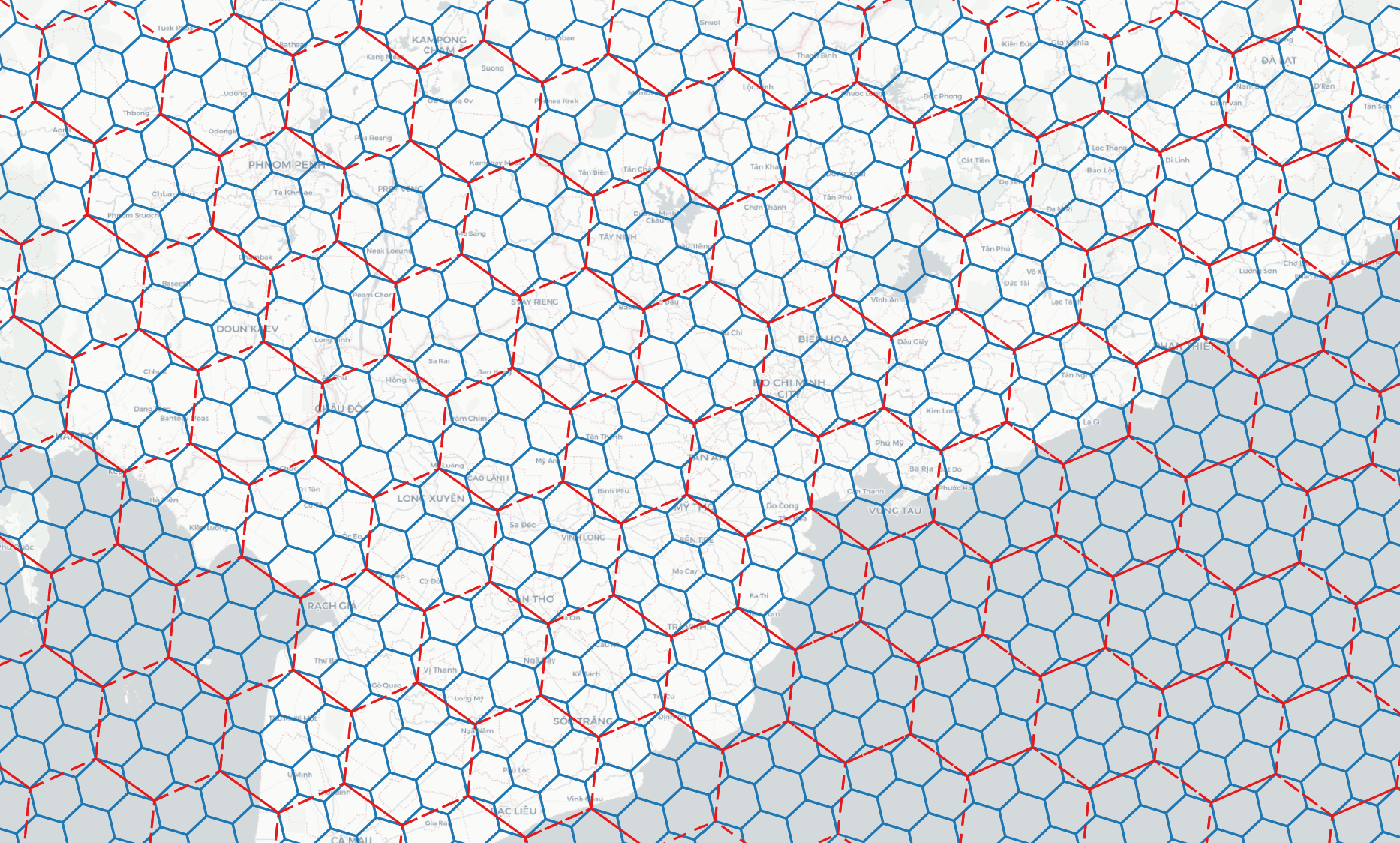
> h3grid -r 11 -b 106.699007 10.762811 106.717674 10.778649 # h3grid -r <resolution> [0..15] -b <min_lon> <min_lat> <max_lon> <max_lat>
> s2grid -r 18 -b 106.699007 10.762811 106.717674 10.778649 # s2grid -r <resolution> [0..30] -b <min_lon> <min_lat> <max_lon> <max_lat>
> rhealpixgrid -r 11 -b 106.699007 10.762811 106.717674 10.778649 # rhealpix2grid -r <resolution> [0..30] -b <min_lon> <min_lat> <max_lon> <max_lat>
> isea4tgrid -r 17 -b 106.699007 10.762811 106.717674 10.778649 # Windows only: isea4tgrid -r <resolution> [0..25] -b <min_lon> <min_lat> <max_lon> <max_lat>
> isea3hgrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # isea3hgrid -r <resolution> [0..32] -b <min_lon> <min_lat> <max_lon> <max_lat>> isea3hstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> easegrid -r 4 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # easegrid -r <resolution> [0..6] -b <min_lon> <min_lat> <max_lon> <max_lat>
> dggridgen -t ISEA3H -r 2 -a ZORDER # Linux only: dggrid -t <DGGS Type> -r <resolution> -a<address_type>.
> qtmgrid -r 8 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # qtmgrid -r <resolution> [1..24] -b <min_lon> <min_lat> <max_lon> <max_lat>
> olcgrid -r 8 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # olcgrid -r <resolution> [2,4,6,8,10..15] -b <min_lon> <min_lat> <max_lon> <max_lat>
> geohashgrid -r 6 -b 106.699007 10.762811 106.717674 10.778649 # geohashgrid -r <resolution> [1..10] -b <min_lon> <min_lat> <max_lon> <max_lat> 1
> geohashgrid -r 2 -b 106.699007 10.762811 106.717674 10.778649 # geohashgrid -r <resolution> [0..5] -b <min_lon> <min_lat> <max_lon> <max_lat>
> georefgrid -r 2 -b 106.699007 10.762811 106.717674 10.778649 # georefgrid -r <resolution> [-1..5] -b <min_lon> <min_lat> <max_lon> <max_lat>
> mgrsgrid -r 1 -gzd 48P # geohashgrid -r <resolution> [0..5] -gzd <Grid Zone Designator, e.g. 48P>
> tilecodegrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # tilecodegrid -r <resolution> [0..26]
> quadkeygrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # quadkeygrid -r <resolution> [0..26]
> maidenheadgrid -r 4 -b 106.699007 10.762811 106.717674 10.778649 # maidenheadgrid -r <resolution> [1..4] -b <min_lon> <min_lat> <max_lon> <max_lat>
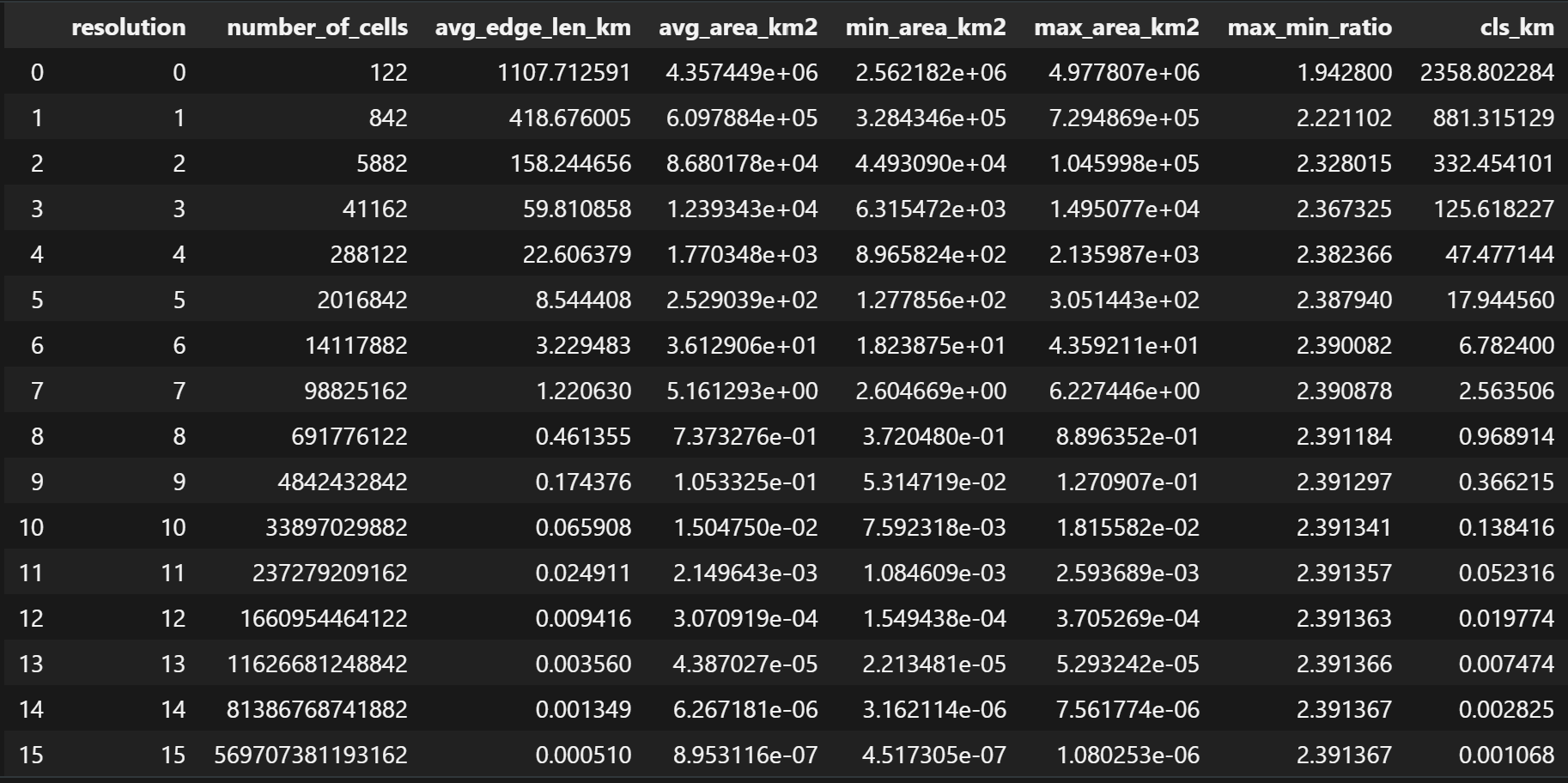
> garsgrid -r 1 -b 106.699007 10.762811 106.717674 10.778649 # garsgrid -r <resolution> [1..4] (30,15,5,1 minutes) -b <min_lon> <min_lat> <max_lon> <max_lat>> h3stats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> s2stats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> rhealpixstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> isea4tstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> isea3hstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> dggridstats -t FULLER3H -r 8 # Linux only: dggrid -t <DGGS Type> -r <resolution>
> easestats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> qtmstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> olcstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> geohashstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> georeftats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> mgrstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> tilecodestats # Number of cells, Cell Width, Cell Height, Cell Area at each resolution
> quadkeystats # Number of cells, Cell Width, Cell Height, Cell Area at each resolution
> maidenheadstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> garsstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution> tetrahedron # Generate Global Tetrahedron
> cube # Generate Global Cube
> octahedron # Generate Global Octahedron
> fuller_icosahedron # Generate Global Fuller Icosahedron
> rhombic_icosahedron # Generate Global rhombic Icosahedron