-
Notifications
You must be signed in to change notification settings - Fork 4
Data included in package
This package contains several example datasets that can be used to explore its functionality. The datasets and simulation scripts used to generate them are contained in their own dedicated branch, and described in detail below.
Once the data have been simulated, it is still incomplete - we must then estimate the individual treatment effects (ITEs) that are used to group observations. The description below also contains information on how the ITEs are estimated and subgroups determined.
- Confounding and no effect modification
- Effect modification and no confounding
- Effect modification and confounding
- Effect modification and confounding by effect modifiers
- Estimation of ITEs and subgroups
The table below defines the possible underlying correlation structures for the covariates in the simple simulated datasets.
-
Y~ N(mu, 1) is the continuous outcome, with the mean given in the table below. -
trt~ Bern(p) is the binary treatment, with the mean given in the table below. -
X5~ N(0,1) is a covariate associated with the treatment only (i.e., an instrument). -
X6~ N(0,1) is a covariate associate with the outcome only (i.e., a prognostic variable). -
X1,X2,X3,X4~i.i.d. N(0,1) are confounders of the effect of the treatment on the outcome. -
E1,E2, andE3~i.i.d. Bern(0.5) are binary effect modifiers that define eight subgroups within the data.
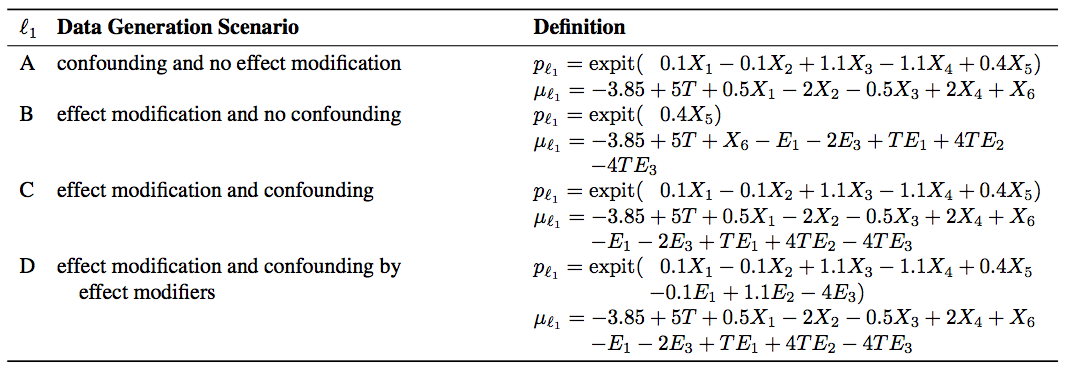
A dataset of size 1500 has been generated under each of the simulation scenarios, and is included upon installation of hetviz. Users are able to generate their own data under this general covariate structure using the function datagen() defined in simpleData.R. This function allows the user to generate a new sample under the population parameters given in the figure above, or specify their own as vectors:
## PSEUDOCODE
# coefficients for the treatment mean
# should be given in the following order, in a
# vector of length 9
alpha <- c(intercept, X1, X2, X3, X4, X5, E1, E2, E3)
# the data were generated using
alpha <- c(0, 0.1, -0.1, 1.1, -1.1, 0.4, -0.1, 1.1, -4)
# coefficients for the outcome mean
# should be given in the following order, in a
# vector of length 13
beta <- c(intercept, trt, X1, X2, X3, X4, X6, E1, E2, E3, TE1, TE2, TE3)
# the data were generated using
beta <- c(-3.85, 5, 0.5, -2, -0.5, 2, 1, -1, 0, -2, 1, 4, -4)Contained in simpleDataA.csv and generated by calling
datagen(n = 1500, effMod = FALSE, confound = TRUE, confoundEMs = FALSE)Contained in simpleDataB.csv and generated by calling
datagen(n = 1500, effMod = TRUE, confound = FALSE, confoundEMs = FALSE)Contained in simpleDataC.csv and generated by calling
datagen(n = 1500, effMod = TRUE, confound = TRUE, confoundEMs = FALSE)Contained in simpleDataD.csv and generated by calling
datagen(n = 1500, effMod = TRUE, confound = TRUE, confoundEMs = TRUE)BayesTree::bart() is used to estimate ITEs. After estimating the ITEs, their quantiles are used to partition the data into subgroups.
An example of how to do is is provided in simpleData.R.
- Medicare data exploring the effect of stent type on revacularization rate